This is a preprint.
Expression, purification, and characterization of diacylated Lipo-YcjN from Escherichia coli
- PMID: 39282304
- PMCID: PMC11398462
- DOI: 10.1101/2024.09.05.611266
Expression, purification, and characterization of diacylated Lipo-YcjN from Escherichia coli
Update in
-
Expression, purification, and characterization of diacylated Lipo-YcjN from Escherichia coli.J Biol Chem. 2024 Nov;300(11):107853. doi: 10.1016/j.jbc.2024.107853. Epub 2024 Oct 1. J Biol Chem. 2024. PMID: 39362470 Free PMC article.
Abstract
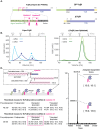
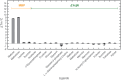
YcjN is a putative substrate-binding protein expressed from a cluster of genes involved in carbohydrate import and metabolism in Escherichia coli. Here, we determine the crystal structure of YcjN to a resolution of 1.95 Å, revealing that its three-dimensional structure is similar to substrate binding proteins in subcluster D-I, which includes the well-characterized maltose binding protein (MBP). Furthermore, we found that recombinant overexpression of YcjN results in the formation of a lipidated form of YcjN that is posttranslationally diacylated at cysteine 21. Comparisons of size-exclusion chromatography profiles and dynamic light scattering measurements of lipidated and non-lipidated YcjN proteins suggest that lipidated YcjN aggregates in solution via its lipid moiety. Additionally, bioinformatic analysis indicates that YcjN-like proteins may exist in both Bacteria and Archaea, potentially in both lipidated and non-lipidated forms. Together, our results provide a better understanding of the aggregation properties of recombinantly expressed bacterial lipoproteins in solution and establish a foundation for future studies that aim to elucidate the role of these proteins in bacterial physiology.
Keywords: Escherichia coli; Substrate binding proteins; X-ray crystallography; bacterial lipoproteins; recombinant protein expression.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
