This is a preprint.
Antigen specificity of clonally-enriched CD8+ T cells in multiple sclerosis
- PMID: 39282370
- PMCID: PMC11398516
- DOI: 10.1101/2024.09.07.611010
Antigen specificity of clonally-enriched CD8+ T cells in multiple sclerosis
Abstract
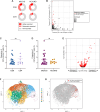
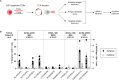
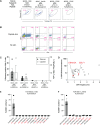
CD8+ T cells are the dominant lymphocyte population in multiple sclerosis (MS) lesions where they are highly clonally expanded. The clonal identity, function, and antigen specificity of CD8+ T cells in MS are not well understood. Here we report a comprehensive single-cell RNA-seq and T cell receptor (TCR)-seq analysis of the cerebrospinal fluid (CSF) and blood from a cohort of treatment-naïve MS patients and control participants. A small subset of highly expanded and activated CD8+ T cells were enriched in the CSF in MS that displayed high activation, cytotoxicity and tissue-homing transcriptional profiles. Using a combination of unbiased and targeted antigen discovery approaches, MS-derived CD8+ T cell clonotypes recognizing Epstein-Barr virus (EBV) antigens and multiple novel mimotopes were identified. These findings shed vital insight into the role of CD8+ T cells in MS and pave the way towards disease biomarkers and therapeutic targets.
Figures


References
-
- Baecher-Allan C., Kaskow B. J. & Weiner H. L. Multiple Sclerosis: Mechanisms and Immunotherapy. Neuron 97, 742–768 (2018). - PubMed
-
- Gay F. W., Drye T. J., Dick G. W. & Esiri M. M. The application of multifactorial cluster analysis in the staging of plaques in early multiple sclerosis. Identification and characterization of the primary demyelinating lesion. Brain 120, 1461–1483 (1997). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
