This is a preprint.
Enhancer activation from transposable elements in extrachromosomal DNA
- PMID: 39282372
- PMCID: PMC11398463
- DOI: 10.1101/2024.09.04.611262
Enhancer activation from transposable elements in extrachromosomal DNA
Update in
-
Enhancer activation from transposable elements in extrachromosomal DNA.Nat Cell Biol. 2025 Nov;27(11):1914-1924. doi: 10.1038/s41556-025-01788-6. Epub 2025 Oct 21. Nat Cell Biol. 2025. PMID: 41120733 Free PMC article.
Abstract
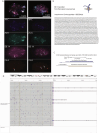
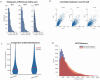
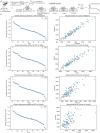
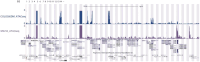
Extrachromosomal DNA (ecDNA) drives oncogene amplification and intratumoral heterogeneity in aggressive cancers. While transposable element (TE) reactivation is common in cancer, its role on ecDNA remains unexplored. Here, we map the 3D architecture of MYC-amplified ecDNA in colorectal cancer cells and identify 68 ecDNA-interacting elements (EIEs)-genomic loci enriched for TEs that are frequently integrated onto ecDNA. We focus on an L1M4a1#LINE/L1 fragment co-amplified with MYC, which functions only in the ecDNA amplified context. Using CRISPR-CATCH, CRISPR interference, and reporter assays, we confirm its presence on ecDNA, enhancer activity, and essentiality for cancer cell fitness. These findings reveal that repetitive elements can be reactivated and co-opted as functional rather than inactive sequences on ecDNA, potentially driving oncogene expression and tumor evolution. Our study uncovers a mechanism by which ecDNA harnesses repetitive elements to shape cancer phenotypes, with implications for diagnosis and therapy.
Conflict of interest statement
Competing Interests H.Y.C. is a cofounder of Accent Therapeutics, Boundless Bio, Cartography Biosciences and Orbital Therapeutics; he was an advisor of 10x Genomics, Arsenal Biosciences, Chroma Medicine and Spring Discovery until 15 December 2024. H.Y.C. is an employee and stockholder of Amgen as of 16 December 2024. M.G.J. is a consultant and holds equity in Tahoe Therapeutics. P.S.M. is a co-founder and advisor of Boundless Bio. J.D.B. is a founder and director of CDI Labs, Inc.; a founder of and consultant to Opentrons LabWorks/Neochromosome, Inc.; and serves or served on the scientific advisory boards of the following: CZ Biohub New York, LLC; Logomix, Inc.; Modern Meadow, Inc.; Rome Therapeutics, Inc.; Sangamo, Inc.; Tessera Therapeutics, Inc.; and the Wyss Institute. V.B. is a cofounder, serves on the scientific advisory board of Boundless Bio and Abterra and holds equity in both companies. Q.S. is an employee and stockholder of Amgen as of 20 February 2025. The remaining authors declare no competing interests.
Figures



References
-
- Yan X., Mischel P. & Chang H. Extrachromosomal DNA in cancer. Nat Rev Cancer 24, 261–273 (2024). - PubMed
-
- Wahl G.M. The importance of circular DNA in mammalian gene amplification. Cancer Res 49, 1333–40 (1989). - PubMed
-
- Benner S.E., Wahl G.M. & Von Hoff D.D. Double minute chromosomes and homogeneously staining regions in tumors taken directly from patients versus in human tumor cell lines. Anticancer Drugs 2, 11–25 (1991). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
