This is a preprint.
Multi-strain phage induced clearance of bacterial infections
- PMID: 39282405
- PMCID: PMC11398464
- DOI: 10.1101/2024.09.07.611814
Multi-strain phage induced clearance of bacterial infections
Update in
-
Multi-strain phage induced clearance of bacterial infections.PLoS Comput Biol. 2025 Feb 4;21(2):e1012793. doi: 10.1371/journal.pcbi.1012793. eCollection 2025 Feb. PLoS Comput Biol. 2025. PMID: 39903766 Free PMC article.
Abstract
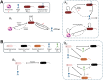
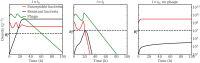
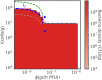
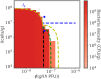
Bacteriophage (or 'phage' - viruses that infect and kill bacteria) are increasingly considered as a therapeutic alternative to treat antibiotic-resistant bacterial infections. However, bacteria can evolve resistance to phage, presenting a significant challenge to the near- and long-term success of phage therapeutics. Application of mixtures of multiple phage (i.e., 'cocktails') have been proposed to limit the emergence of phage-resistant bacterial mutants that could lead to therapeutic failure. Here, we combine theory and computational models of in vivo phage therapy to study the efficacy of a phage cocktail, composed of two complementary phages motivated by the example of Pseudomonas aeruginosa facing two phages that exploit different surface receptors, LUZ19v and PAK_P1. As confirmed in a Luria-Delbrück fluctuation test, this motivating example serves as a model for instances where bacteria are extremely unlikely to develop simultaneous resistance mutations against both phages. We then quantify therapeutic outcomes given single- or double-phage treatment models, as a function of phage traits and host immune strength. Building upon prior work showing monophage therapy efficacy in immunocompetent hosts, here we show that phage cocktails comprised of phage targeting independent bacterial receptors can improve treatment outcome in immunocompromised hosts and reduce the chance that pathogens simultaneously evolve resistance against phage combinations. The finding of phage cocktail efficacy is qualitatively robust to differences in virus-bacteria interactions and host immune dynamics. Altogether, the combined use of theory and computational analysis highlights the influence of viral life history traits and receptor complementarity when designing and deploying phage cocktails in immunocompetent and immunocompromised hosts.
Figures


References
-
- Suttle CA (2007) Marine viruses — major players in the global ecosystem. Nature Reviews Microbiology 5:801–812. - PubMed
-
- Labrie SJ, Samson JE, Moineau S (2010) Bacteriophage resistance mechanisms. Nature Reviews Microbiology 8:317–327. - PubMed
-
- Hampton HG, Watson BNJ, Fineran PC (2020) The arms race between bacteria and their phage foes. Nature 577:327–336. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
