This is a preprint.
Examining intra-host genetic variation of RSV by short read high-throughput sequencing
- PMID: 39282457
- PMCID: PMC11398394
- DOI: 10.1101/2023.05.17.541198
Examining intra-host genetic variation of RSV by short read high-throughput sequencing
Abstract
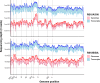
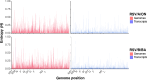
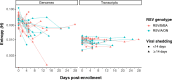
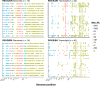
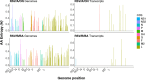
Every viral infection entails an evolving population of viral genomes. High-throughput sequencing technologies can be used to characterize such populations, but to date there are few published examples of such work. In addition, mixed sequencing data are sometimes used to infer properties of infecting genomes without discriminating between genome-derived reads and reads from the much more abundant, in the case of a typical active viral infection, transcripts. Here we apply capture probe-based short read high-throughput sequencing to nasal wash samples taken from a previously described group of adult hematopoietic cell transplant (HCT) recipients naturally infected with respiratory syncytial virus (RSV). We separately analyzed reads from genomes and transcripts for the levels and distribution of genetic variation by calculating per position Shannon entropies. Our analysis reveals a low level of genetic variation within the RSV infections analyzed here, but with interesting differences between genomes and transcripts in 1) average per sample Shannon entropies; 2) the genomic distribution of variation 'hotspots'; and 3) the genomic distribution of hotspots encoding alternative amino acids. In all, our results suggest the importance of separately analyzing reads from genomes and transcripts when interpreting high-throughput sequencing data for insight into intra-host viral genome replication, expression, and evolution.
Figures

References
-
- Nelson MI, Simonsen L, Viboud C, Miller MA, Taylor J, George KS, Griesemer SB, Ghedin E, Sengamalay NA, Spiro DJ, Volkov I, Grenfell BT, Lipman DJ, Taubenberger JK, Holmes EC. 2006. Stochastic processes are key determinants of short-term evolution in influenza a virus. PLoS Pathog 2:e125. - PMC - PubMed
-
- Avadhanula V, Chemaly RF, Shah DP, Ghantoji SS, Azzi JM, Aideyan LO, Mei M, Piedra PA. 2015. Infection with novel respiratory syncytial virus genotype Ontario (ON1) in adult hematopoietic cell transplant recipients, Texas, 2011–2013. J Infect Dis 211:582–589. - PubMed
-
- Ye X, Cabral de Rezende W, Iwuchukwu OP, Avadhanula V, Ferlic-Stark LL, Patel KD, Piedra FA, Shah DP, Chemaly RF, Piedra PA. 2020. Antibody Response to the Furin Cleavable Twenty-Seven Amino Acid Peptide (p27) of the Fusion Protein in Respiratory Syncytial Virus (RSV) Infected Adult Hematopoietic Cell Transplant (HCT) Recipients. Vaccines (Basel) 8. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
