Different molecular recognition by three domains of the full-length GRB2 to SOS1 proline-rich motifs and EGFR phosphorylated sites
- PMID: 39282643
- PMCID: PMC11391413
- DOI: 10.1039/d4sc02656j
Different molecular recognition by three domains of the full-length GRB2 to SOS1 proline-rich motifs and EGFR phosphorylated sites
Abstract
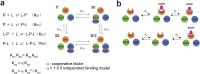
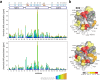
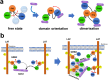
The adaptor protein human GRB2 plays crucial roles in mediating signal transduction from cell membrane receptors to RAS and its downstream proteins by recruiting SOS1. Recent studies have revealed that GRB2 also serves as a scaffold for liquid-liquid phase separation (LLPS) with SOS1 and transmembrane receptors, which is thought to regulate the magnitude of cell signalling pathways. In this study, we employed solution NMR spectroscopy to investigate the interactions of the full-length GRB2 with proline-rich motifs (PRMs) derived from ten potential GRB2-binding sites in SOS1, as well as a peptide from a phosphorylation site of EGFR. Our findings indicate that the binding affinity of the two SH3 domains of GRB2 for PRMs differs by a factor of ten to twenty, with the N-terminal SH3 domain (NSH3) exhibiting a markedly higher affinity. The interactions of PRMs with the SH3 domains affected not only the regions surrounding the PRM binding sites on the SH3 domains but also the linker area connecting the three domains and parts of the SH2 domain. Analysis of the interaction between the phosphorylated EGFR binding site and the SH2 domain revealed chemical shift perturbations in regions distal from the known binding site of SH2. Moreover, we observed that the inter-domain interactions of the two SH3 domains with the SH2 domain of GRB2 are asymmetric. These findings suggest that the local binding of PRMs and phosphorylated EGFR to GRB2 impacts the overall structure of the GRB2 molecule, including domain orientation and dimerisation, which may contribute to LLPS formation.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

