Identifying Candida auris transmission in a hospital outbreak investigation using whole-genome sequencing and SNP phylogenetic analysis
- PMID: 39283080
- PMCID: PMC11481579
- DOI: 10.1128/jcm.00680-24
Identifying Candida auris transmission in a hospital outbreak investigation using whole-genome sequencing and SNP phylogenetic analysis
Abstract
Candida auris poses a global public health challenge, causing multiple outbreaks within healthcare facilities. Despite advancements in strain typing for various infectious diseases, a consensus on the genetic relatedness threshold for identifying C. auris transmission in local hospital outbreaks remains elusive. We investigated genetic variations within our local isolate collection using whole-genome-based single nucleotide polymorphism (SNP) phylogenetic analysis. A total of 74 C. auris isolates were subjected to whole-genome sequencing (WGS) and SNP phylogenetic analysis via the QIAGEN CLC Genomics Workbench. Isolates included known related strains from the same patient, strains from different hospitals, strains from our hospital patients with no epidemiological link, and 19 patient isolates from a recent C. auris outbreak. All but three isolates were identified to be Clade IV. By examining the genetic diversities of C. auris within patients and between patients, we identified a SNP variation range of 0-13 for identifying related isolates. During an outbreak investigation, utilizing this range, maximum likelihood phylogenetic analysis revealed two distinct clusters that aligned with the epidemiological links. Determining a SNP variation range to delineate genetic relatedness among isolates is crucial for the application of WGS and SNP phylogenetic analysis in identifying C. auris transmission during hospital outbreak investigations. The use of WGS SNP phylogenetic analysis via the CLC Genomics Workbench has emerged as a valuable method for typing C. auris in clinical microbiology laboratories.
Keywords: Candida auris; WG-SNP; anti-fungal susceptibilities; outbreak; strain typing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
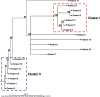
References
-
- Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, et al. 2013. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus — United States, May 2013–August 2016. MMWR Morb Mortal Wkly Rep 65:1234–1237. doi: 10.15585/mmwr.mm6544e1 - DOI - PubMed
-
- Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

