Bacteriocin distribution patterns in Enterococcus faecium and Enterococcus lactis: bioinformatic analysis using a tailored genomics framework
- PMID: 39283104
- PMCID: PMC11497781
- DOI: 10.1128/aem.01376-24
Bacteriocin distribution patterns in Enterococcus faecium and Enterococcus lactis: bioinformatic analysis using a tailored genomics framework
Abstract
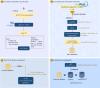
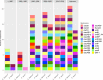
Multidrug-resistant Enterococcus faecium strains represent a major concern due to their ability to thrive in diverse environments and cause life-threatening infections. While antimicrobial resistance and virulence mechanisms have been extensively studied, the contribution of bacteriocins to E. faecium's adaptability remains poorly explored. E. faecium, within the Bacillota phylum, is a prominent bacteriocin producer. Here, we developed a tailored database of 76 Bacillota bacteriocins (217 sequences, including 40 novel bacteriocins) and applied it to uncover bacteriocin distribution patterns in 997 quality-filtered E. faecium and Enterococcus lactis (former E. faecium clade B) genomes. Curated using computational pipelines and literature mining, our database demonstrates superior precision versus leading public tools in identifying diverse bacteriocins. Distinct bacteriocin profiles emerged between E. faecium and E. lactis, highlighting species-specific adaptations. E. faecium strains from hospitalized patients were significantly enriched in bacteriocins as enterocin A and bacteriocins 43 (or T8), AS5, and AS11. These bacteriocin genes were strongly associated with antibiotic resistance, particularly vancomycin and ampicillin, and Inc18 rep2_pRE25-derivative plasmids, classically associated with vancomycin resistance transposons. Such bacteriocin arsenal likely enhances the adaptability and competitive fitness of E. faecium in the nosocomial environment. By combining a novel tailored database, whole-genome sequencing, and epidemiological data, our work elucidates meaningful connections between bacteriocin determinants, antimicrobial resistance, mobile genetic elements, and ecological origins in E. faecium and provides a framework for elucidating bacteriocin landscapes in other organisms. Characterizing species- and strain-level differences in bacteriocin profiles may reveal determinants of ecological adaptation, and translating these discoveries could further inform strategies to exploit bacteriocins against high-risk clones.
Importance: This work significantly expands the knowledge on the understudied bacteriocin diversity in opportunistic enterococci, revealing their contribution in the adaptation to different environments. It underscores the importance of placing increased emphasis on genetic platforms carrying bacteriocins as well as on cryptic plasmids that often exclusively harbor bacteriocins since bacteriocin production can significantly contribute to plasmid maintenance, potentially facilitating their stable transmission across generations. Further characterization of strain-level bacteriocin landscapes could inform strategies to combat high-risk clones. Overall, these insights provide a framework for unraveling the therapeutic and biotechnological potential of bacteriocins.
Keywords: Enterococcus faecium; Enterococcus lactis; antimicrobial peptides; antimicrobial resistance; bacteriocin database; bacteriocins; plasmids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Ness IF, Diep DB, Ike Y. 2014. Enterococcal bacteriocins and antimicrobial proteins that contribute to niche control. In Gilmore MS, Clewell DB, Ike Y, Shankar N (ed), Enterococci: from commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

