Evolution recovers the fitness of Acinetobacter baylyi strains with large deletions through mutations in deletion-specific targets and global post-transcriptional regulators
- PMID: 39283914
- PMCID: PMC11426457
- DOI: 10.1371/journal.pgen.1011306
Evolution recovers the fitness of Acinetobacter baylyi strains with large deletions through mutations in deletion-specific targets and global post-transcriptional regulators
Abstract
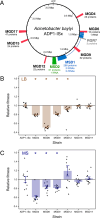
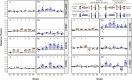
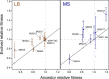
Organelles and endosymbionts have naturally evolved dramatically reduced genome sizes compared to their free-living ancestors. Synthetic biologists have purposefully engineered streamlined microbial genomes to create more efficient cellular chassis and define the minimal components of cellular life. During natural or engineered genome streamlining, deletion of many non-essential genes in combination often reduces bacterial fitness for idiosyncratic or unknown reasons. We investigated how and to what extent laboratory evolution could overcome these defects in six variants of the transposon-free Acinetobacter baylyi strain ADP1-ISx that each had a deletion of a different 22- to 42-kilobase region and two strains with larger deletions of 70 and 293 kilobases. We evolved replicate populations of ADP1-ISx and each deletion strain for ~300 generations in a chemically defined minimal medium or a complex medium and sequenced the genomes of endpoint clonal isolates. Fitness increased in all cases that were examined except for two ancestors that each failed to improve in one of the two environments. Mutations affecting nine protein-coding genes and two small RNAs were significantly associated with one of the two environments or with certain deletion ancestors. The global post-transcriptional regulators rnd (ribonuclease D), csrA (RNA-binding carbon storage regulator), and hfq (RNA-binding protein and chaperone) were frequently mutated across all strains, though the incidence and effects of these mutations on gene function and bacterial fitness varied with the ancestral deletion and evolution environment. Mutations in this regulatory network likely compensate for how an earlier deletion of a transposon in the ADP1-ISx ancestor of all the deletion strains restored csrA function. More generally, our results demonstrate that fitness lost during genome streamlining can usually be regained rapidly through laboratory evolution and that recovery tends to occur through a combination of deletion-specific compensation and global regulatory adjustments.
Copyright: © 2024 Gifford et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

