Brain tumor segmentation by combining MultiEncoder UNet with wavelet fusion
- PMID: 39284311
- PMCID: PMC11540057
- DOI: 10.1002/acm2.14527
Brain tumor segmentation by combining MultiEncoder UNet with wavelet fusion
Abstract
Background and objective: Accurate segmentation of brain tumors from multimodal magnetic resonance imaging (MRI) holds significant importance in clinical diagnosis and surgical intervention, while current deep learning methods cope with situations of multimodal MRI by an early fusion strategy that implicitly assumes that the modal relationships are linear, which tends to ignore the complementary information between modalities, negatively impacting the model's performance. Meanwhile, long-range relationships between voxels cannot be captured due to the localized character of the convolution procedure.
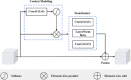
Method: Aiming at this problem, we propose a multimodal segmentation network based on a late fusion strategy that employs multiple encoders and a decoder for the segmentation of brain tumors. Each encoder is specialized for processing distinct modalities. Notably, our framework includes a feature fusion module based on a 3D discrete wavelet transform aimed at extracting complementary features among the encoders. Additionally, a 3D global context-aware module was introduced to capture the long-range dependencies of tumor voxels at a high level of features. The decoder combines fused and global features to enhance the network's segmentation performance.
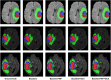
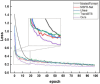
Result: Our proposed model is experimented on the publicly available BraTS2018 and BraTS2021 datasets. The experimental results show competitiveness with state-of-the-art methods.
Conclusion: The results demonstrate that our approach applies a novel concept for multimodal fusion within deep neural networks and delivers more accurate and promising brain tumor segmentation, with the potential to assist physicians in diagnosis.
Keywords: 3D discrete wavelet transformer; brain tumor segmentation; multi‐encoder.
© 2024 The Author(s). Journal of Applied Clinical Medical Physics published by Wiley Periodicals LLC on behalf of American Association of Physicists in Medicine.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







References
-
- Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35(5):1240‐1251. - PubMed
-
- Choi SG, Sohn CB. Detection of HGG and LGG brain tumors using U‐Net. Medico‐legal Update. 2019;19(1):560‐565.
-
- Shukla G, Alexander GS, Bakas S, et al. Advanced magnetic resonance imaging in glioblastoma: a review. Chin Clin Oncol. 2017;6(4):40. - PubMed
-
- Bakas S, Reyes M, Jakab A, et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. [published online ahead of print April 23, 2019]. Computer Vision and Pattern Recognition. doi:10.48550/arXiv.1811.02629 - DOI
-
- Yue W, Wang Z, Tian B, Pook M, Liu X. A hybrid model‐and memory‐based collaborative filtering algorithm for baseline data prediction of Friedreich's ataxia patients. IEEE Trans Ind Inf. 2020;17(2):1428‐1437.
MeSH terms
Grants and funding
- 62204168/National Natural Science Foundation of China
- 20YDTPJC00160/Tianjin Municipal Science and Technology Program
- 21YDTPJC00780/Tianjin Municipal Science and Technology Program
- 2019KJ101/Science Research Program of Tianjin Education Committee
- 2022KYZ136/Tianjin Research Innovation Project for Postgraduate Students
LinkOut - more resources
Full Text Sources
Medical

