Unraveling the Genetics of Shared Clinical and Serological Manifestations in Patients With Systemic Inflammatory Autoimmune Diseases
- PMID: 39284741
- PMCID: PMC11782108
- DOI: 10.1002/art.42988
Unraveling the Genetics of Shared Clinical and Serological Manifestations in Patients With Systemic Inflammatory Autoimmune Diseases
Abstract
Objective: Systemic inflammatory autoimmune diseases (SIADs) such as systemic lupus erythematosus (SLE), primary Sjögren disease (pSS), and idiopathic inflammatory myopathies (myositis) are complex conditions characterized by shared circulating autoantibodies and clinical manifestations, including skin rashes, among others. This study was aimed at elucidating the genetics underlying these common features.
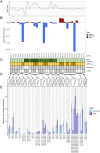
Methods: We performed targeted DNA sequencing of coding and regulatory regions from approximately 1,900 immune-related genes in a large cohort of 2,292 well-characterized Scandinavian patients with SIADs with SLE, pSS, and myositis as well as 1,252 controls. A gene-based functionally weighted genetic score for aggregate testing of all genetic variants, including rare variants, was complemented by in silico functional analyses and in vitro reporter experiments.
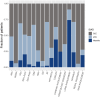
Results: Case-control association analysis detected known and potentially novel genetic loci in agreement with previous genetic and transcriptomics findings linked to the SIAD autoimmune background. Intriguingly, case-case comparisons between patient subgroups with and without specific autoantibodies revealed that the subgroups defined by antinuclear antibodies and anti-double-stranded DNA antibodies have unique genetic profiles reflecting their heterogeneity. When focusing on clinical features, we overall showed that dual-specificity phosphatase 1 (DUSP1) protective genetic variants lead to increased gene expression and potentially to anti-inflammatory effects on the SIAD-associated skin phenotype. This is consistent with recent genetic findings on eczema and with the previously reported down-regulation of the MAPK signaling-related gene DUSP1 in other skin disorders.
Conclusion: Together, this suggests common molecular mechanisms potentially underlying overlapping clinical manifestations shared among different disorders and informs clinical heterogeneity, which could be translated to improve disease diagnostic and treatment, also in more generalized disease frameworks.
© 2024 The Author(s). Arthritis & Rheumatology published by Wiley Periodicals LLC on behalf of American College of Rheumatology.
Figures



References
-
- Bengtsson AA, Ronnblom L. Systemic lupus erythematosus: still a challenge for physicians. J Intern Med 2017;281(1):52–64. - PubMed
-
- Dalakas MC. Inflammatory muscle diseases. N Engl J Med 2015;372(18):1734–1747. - PubMed
-
- Mariette X, Criswell LA. Primary Sjogren's syndrome. N Engl J Med 2018;378(10):931–939. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

