Lipoprotein metabolism mediates hematopoietic stem cell responses under acute anemic conditions
- PMID: 39284836
- PMCID: PMC11405780
- DOI: 10.1038/s41467-024-52509-w
Lipoprotein metabolism mediates hematopoietic stem cell responses under acute anemic conditions
Abstract
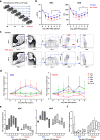
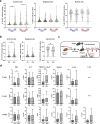
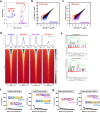
Hematopoietic stem cells (HSCs) react to various stress conditions. However, it is unclear whether and how HSCs respond to severe anemia. Here, we demonstrate that upon induction of acute anemia, HSCs rapidly proliferate and enhance their erythroid differentiation potential. In severe anemia, lipoprotein profiles largely change and the concentration of ApoE increases. In HSCs, transcription levels of lipid metabolism-related genes, such as very low-density lipoprotein receptor (Vldlr), are upregulated. Stimulation of HSCs with ApoE enhances their erythroid potential, whereas HSCs in Apoe knockout mice do not respond to anemia induction. VldlrhighHSCs show higher erythroid potential, which is enhanced after acute anemia induction. VldlrhighHSCs are epigenetically distinct because of their low chromatin accessibility, and more chromatin regions are closed upon acute anemia induction. Chromatin regions closed upon acute anemia induction are mainly binding sites of Erg. Inhibition of Erg enhanced the erythroid differentiation potential of HSCs. Our findings indicate that lipoprotein metabolism plays an important role in HSC regulation under severe anemic conditions.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

