RNA fine-tunes estrogen receptor-alpha binding on low-affinity DNA motifs for transcriptional regulation
- PMID: 39284910
- PMCID: PMC11535219
- DOI: 10.1038/s44318-024-00225-y
RNA fine-tunes estrogen receptor-alpha binding on low-affinity DNA motifs for transcriptional regulation
Abstract
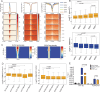
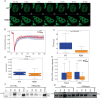
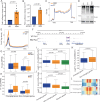
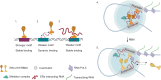
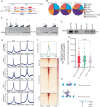
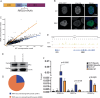
Transcription factors (TFs) regulate gene expression by binding with varying strengths to DNA via their DNA-binding domain. Additionally, some TFs also interact with RNA, which modulates transcription factor binding to chromatin. However, whether RNA-mediated TF binding results in differential transcriptional outcomes remains unknown. In this study, we demonstrate that estrogen receptor α (ERα), a ligand-activated TF, interacts with RNA in a ligand-dependent manner. Defects in RNA binding lead to genome-wide loss of ERα recruitment, particularly at weaker ERα-motifs. Furthermore, ERα mobility in the nucleus increases in the absence of its RNA-binding capacity. Unexpectedly, this increased mobility coincides with robust polymerase loading and transcription of ERα-regulated genes that harbor low-strength motifs. However, highly stable binding of ERα on chromatin negatively impacts ligand-dependent transcription. Collectively, our results suggest that RNA interactions spatially confine ERα on low-affinity sites to fine-tune gene transcription.
Keywords: Chromatin; DNA-motifs; Estrogen Receptor-Alpha; Non-Coding RNA; Transcription Factors.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Anon. n.d. Picard tools - by Broad Institute. http://broadinstitute.github.io/picard/
-
- Aiello A, Bacci L, Re A, Ripoli C, Pierconti F, Pinto F, Masetti R, Grassi C, Gaetano C, Bassi PF, Pontecorvi A, Nanni S, Farsetti A (2016) MALAT1 and HOTAIR long non-coding RNAs play opposite role in estrogen-mediated transcriptional regulation in prostate cancer cells. Sci Rep. 6:38414 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

