7D, a small molecule inhibits dengue infection by increasing interferons and neutralizing-antibodies via CXCL4:CXCR3:p38:IRF3 and Sirt1:STAT3 axes respectively
- PMID: 39284947
- PMCID: PMC11473809
- DOI: 10.1038/s44321-024-00137-8
7D, a small molecule inhibits dengue infection by increasing interferons and neutralizing-antibodies via CXCL4:CXCR3:p38:IRF3 and Sirt1:STAT3 axes respectively
Abstract
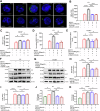
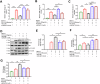
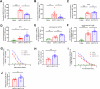
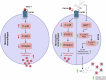
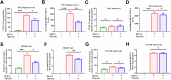
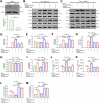
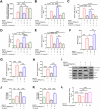
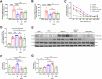
There are a limited number of effective vaccines against dengue virus (DENV) and significant efforts are being made to develop potent anti-virals. Previously, we described that platelet-chemokine CXCL4 negatively regulates interferon (IFN)-α/β synthesis and promotes DENV2 replication. An antagonist to CXCR3 (CXCL4 receptor) reversed it and inhibited viral replication. In a concurrent search, we identified CXCR3-antagonist from our compound library, namely 7D, which inhibited all serotypes of DENV in vitro. With a half-life of ~2.85 h in plasma and no significant toxicity, 7D supplementation (8 mg/kg-body-weight) to DENV2-infected IFNα/β/γR-/-AG129 or wild-type C57BL6 mice increased synthesis of IFN-α/β and IFN-λ, and rescued disease symptoms like thrombocytopenia, leukopenia and vascular-leakage, with improved survival. 7D, having the property to inhibit Sirt-1 deacetylase, promoted acetylation and phosphorylation of STAT3, which in-turn increased plasmablast proliferation, germinal-center maturation and synthesis of neutralizing-antibodies against DENV2 in mice. A STAT3-inhibitor successfully inhibited these effects of 7D. Together, these observations identify compound 7D as a stimulator of IFN-α/β/λ synthesis via CXCL4:CXCR3:p38:IRF3 signaling, and a booster for neutralizing-antibody generation by promoting STAT3-acetylation in plasmablasts, capable of protecting dengue infection.
Keywords: Antibodies; CXCL4; CXCR3-antagonist; Dengue; Interferons.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Case DA, Aktulga HM, Belfon K, Ben-Shalom IY, Berryman JT, Brozell SR, Cerutti DS, Cheatham TE III, Cisneros GA, Cruzeiro VWD et al (2022) AMBER 22 reference manual. University of California, San Francisco, CA
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

