Neoadjuvant nivolumab or nivolumab plus ipilimumab in early-stage triple-negative breast cancer: a phase 2 adaptive trial
- PMID: 39284953
- PMCID: PMC11564107
- DOI: 10.1038/s41591-024-03249-3
Neoadjuvant nivolumab or nivolumab plus ipilimumab in early-stage triple-negative breast cancer: a phase 2 adaptive trial
Abstract
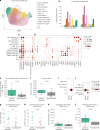
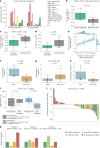
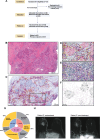
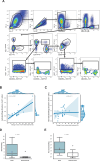
Immune checkpoint inhibition (ICI) with chemotherapy is now the standard of care for stage II-III triple-negative breast cancer; however, it is largely unknown for which patients ICI without chemotherapy could be an option and what the benefit of combination ICI could be. The adaptive BELLINI trial explored whether short combination ICI induces immune activation (primary end point, twofold increase in CD8+ T cells or IFNG), providing a rationale for neoadjuvant ICI without chemotherapy. Here, in window-of-opportunity cohorts A (4 weeks of anti-PD-1) and B (4 weeks of anti-PD-1 + anti-CTLA4), we observed immune activation in 53% (8 of 15) and 60% (9 of 15) of patients, respectively. High levels of tumor-infiltrating lymphocytes correlated with response. Single-cell RNA sequencing revealed that higher pretreatment tumor-reactive CD8+ T cells, follicular helper T cells and shorter distances between tumor and CD8+ T cells correlated with response. Higher levels of regulatory T cells after treatment were associated with nonresponse. Based on these data, we opened cohort C for patients with high levels of tumor-infiltrating lymphocytes (≥50%) who received 6 weeks of neoadjuvant anti-PD-1 + anti-CTLA4 followed by surgery (primary end point, pathological complete response). Overall, 53% (8 of 15) of patients had a major pathological response (<10% viable tumor) at resection, with 33% (5 of 15) having a pathological complete response. All cohorts met Simon's two-stage threshold for expansion to stage II. We observed grade ≥3 adverse events for 17% of patients and a high rate (57%) of immune-mediated endocrinopathies. In conclusion, neoadjuvant immunotherapy without chemotherapy demonstrates potential efficacy and warrants further investigation in patients with early triple-negative breast cancer. ClinicalTrials.gov registration: NCT03815890 .
© 2024. The Author(s).
Conflict of interest statement
Figures





References
-
- Schmid, P. et al. Event-free survival with pembrolizumab in early triple-negative breast cancer. N. Engl. J. Med.386, 556–567 (2022). - PubMed
-
- Loibl, S. et al. A randomised phase II study investigating durvalumab in addition to an anthracycline taxane-based neoadjuvant therapy in early triple-negative breast cancer: clinical results and biomarker analysis of GeparNuevo study. Ann. Oncol.30, 1279–1288 (2019). - PubMed
-
- Mittendorf, E. A. et al. Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): a randomised, double-blind, phase 3 trial. Lancet396, 1090–1100 (2020). - PubMed
-
- Schmid, P. et al. Pembrolizumab for early triple-negative breast cancer. N. Engl. J. Med.382, 810–821 (2020). - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

