ctDNA-based molecular residual disease and survival in resectable colorectal cancer
- PMID: 39284954
- PMCID: PMC11564113
- DOI: 10.1038/s41591-024-03254-6
ctDNA-based molecular residual disease and survival in resectable colorectal cancer
Abstract
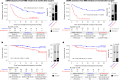
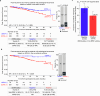
The interim analysis of the CIRCULATE-Japan GALAXY observational study demonstrated the association of circulating tumor DNA (ctDNA)-based molecular residual disease (MRD) detection with recurrence risk and benefit from adjuvant chemotherapy (ACT) in resectable colorectal cancer (CRC). This updated analysis with a 23-month median follow-up, including 2,240 patients with stage II-III colon cancer or stage IV CRC, reinforces the prognostic value of ctDNA positivity during the MRD window with significantly inferior disease-free survival (DFS; hazard ratio (HR): 11.99, P < 0.0001) and overall survival (OS; HR: 9.68, P < 0.0001). In patients who experienced recurrence, ctDNA positivity correlated with shorter OS (HR: 2.71, P < 0.0001). The significantly shorter DFS in MRD-positive patients was consistent across actionable biomarker subsets. Sustained ctDNA clearance in response to ACT was an indicator of favorable DFS and OS compared to transient clearance (24-month DFS: 89.0% versus 3.3%; 24-month OS: 100.0% versus 82.3%). True spontaneous clearance rate with no clinical recurrence was 1.9% (2/105). Overall, our findings provide evidence for the utility of ctDNA monitoring for post-resection recurrence and mortality risk stratification that could be used for guiding adjuvant therapy.
© 2024. The Author(s).
Conflict of interest statement
Figures













References
-
- Kasi, P. M. et al. Circulating tumor DNA (ctDNA) for informing adjuvant chemotherapy (ACT) in stage II/III colorectal cancer (CRC): interim analysis of BESPOKE CRC study. J. Clin. Oncol. 42, 10.1200/JCO.2024.42.3_suppl.9 (2024).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

