Agnostic B cell selection approach identifies antibodies against K. pneumoniae that synergistically drive complement activation
- PMID: 39285158
- PMCID: PMC11405761
- DOI: 10.1038/s41467-024-52372-9
Agnostic B cell selection approach identifies antibodies against K. pneumoniae that synergistically drive complement activation
Abstract
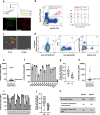
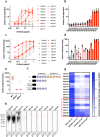
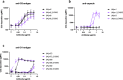
Antibody-dependent complement activation plays a key role in the natural human immune response to infections. Currently, the understanding of which antibody-antigen combinations drive a potent complement response on bacteria is limited. Here, we develop an antigen-agnostic approach to stain and single-cell sort human IgG memory B cells recognizing intact bacterial cells, keeping surface antigens in their natural context. With this method we successfully identified 29 antibodies against K. pneumoniae, a dominant cause of hospital-acquired infections with increasing antibiotic resistance. Combining genetic tools and functional analyses, we reveal that the capacity of antibodies to activate complement on K. pneumoniae critically depends on their antigenic target. Furthermore, we find that antibody combinations can synergistically activate complement on K. pneumoniae by strengthening each other's binding in an Fc-independent manner. Understanding the molecular basis of effective complement activation by antibody combinations to mimic a polyclonal response could accelerate the development of antibody-based therapies against problematic infections.
© 2024. The Author(s).
Conflict of interest statement
R.J.M., J.S., and F.J.B. are (former) employees at Genmab BV and have ownership interests (including stocks, warrants, patents, etc.). Other authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- 16442/Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Netherlands Organisation for Scientific Research)
- 101001937/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources

