Design of fully synthetic signal peptide library and its use for enhanced secretory production of recombinant proteins in Corynebacterium glutamicum
- PMID: 39285401
- PMCID: PMC11406804
- DOI: 10.1186/s12934-024-02516-9
Design of fully synthetic signal peptide library and its use for enhanced secretory production of recombinant proteins in Corynebacterium glutamicum
Abstract
Background: Corynebacterium glutamicum is an attractive host for secretory production of recombinant proteins, including high-value industrial enzymes and therapeutic proteins. The choice of an appropriate signaling peptide is crucial for efficient protein secretion. However, due to the limited availability of signal peptides in C. glutamicum, establishing an optimal secretion system is challenging.
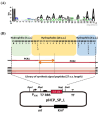
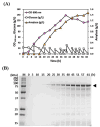
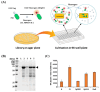
Result: We constructed a signal peptide library for the isolation of target-specific signal peptides and developed a highly efficient secretory production system in C. glutamicum. Based on the sequence information of the signal peptides of the general secretion-dependent pathway in C. glutamicum, a synthetic signal peptide library was designed, and validated with three protein models. First, we examined endoxylanase (XynA) and one potential signal peptide (C1) was successfully isolated by library screening on xylan-containing agar plates. With this C1 signal peptide, secretory production of XynA as high as 3.2 g/L could be achieved with high purity (> 80%). Next, the signal peptide for ⍺-amylase (AmyA) was screened on a starch-containing agar plate. The production titer of the isolated signal peptide (HS06) reached 1.48 g/L which was 2-fold higher than that of the well-known Cg1514 signal peptide. Finally, we isolated the signal peptide for the M18 single-chain variable fragment (scFv). As an enzyme-independent screening tool, we developed a fluorescence-dependent screening tool using Fluorescence-Activating and Absorption-Shifting Tag (FAST) fusion, and successfully isolated the optimal signal peptide (18F11) for M18 scFv. With 18F11, secretory production as high as 228 mg/L was achieved, which was 3.4-fold higher than previous results.
Conclusions: By screening a fully synthetic signal peptide library, we achieved improved production of target proteins compared to previous results using well-known signal peptides. Our synthetic library provides a useful resource for the development of an optimal secretion system for various recombinant proteins in C. glutamicum, and we believe this bacterium to be a more promising workhorse for the bioindustry.
Keywords: Corynebacterium glutamicum; Library screening; Sec-dependent pathway; Secretion; Synthetic signal peptide.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Lee SM, Jeong KJ. Advances in synthetic biology tools and engineering of Corynebacterium glutamicum as a platform host for recombinant protein production. Biotechnol Bioproc E. 2023;28(6):962–76. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

