Whole-genome sequencing of major malaria vectors reveals the evolution of new insecticide resistance variants in a longitudinal study in Burkina Faso
- PMID: 39285410
- PMCID: PMC11406867
- DOI: 10.1186/s12936-024-05106-7
Whole-genome sequencing of major malaria vectors reveals the evolution of new insecticide resistance variants in a longitudinal study in Burkina Faso
Abstract
Background: Intensive deployment of insecticide based malaria vector control tools resulted in the rapid evolution of phenotypes resistant to these chemicals. Understanding this process at the genomic level is important for the deployment of successful vector control interventions. Therefore, longitudinal sampling followed by whole genome sequencing (WGS) is necessary to understand how these evolutionary processes evolve over time. This study investigated the change in genetic structure and the evolution of the insecticide resistance variants in natural populations of Anopheles gambiae over time and space from 2012 to 2017 in Burkina Faso.
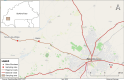
Methods: New genomic data have been generated from An. gambiae mosquitoes collected from three villages in the western part of Burkina Faso between 2012 and 2017. The samples were whole-genome sequenced and the data used in the An. gambiae 1000 genomes (Ag1000G) project as part of the Vector Observatory. Genomic data were analysed using the analysis pipeline previously designed by the Ag1000G project.
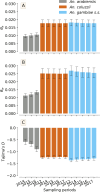
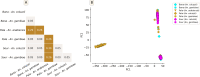
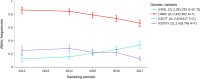
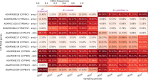
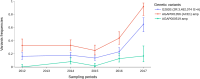
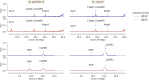
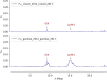
Results: The results showed similar and consistent nucleotide diversity and negative Tajima's D between An. gambiae sensu stricto (s.s.) and Anopheles coluzzii. Principal component analysis (PCA) and the fixation index (FST) showed a clear genetic structure in the An. gambiae sensu lato (s.l.) species. Genome-wide FST and H12 scans identified genomic regions under divergent selection that may have implications in the adaptation to ecological changes. Novel voltage-gated sodium channel pyrethroid resistance target-site alleles (V402L, I1527T) were identified at increasing frequencies alongside the established alleles (Vgsc-L995F, Vgsc-L995S and N1570Y) within the An. gambiae s.l.
Populations: Organophosphate metabolic resistance markers were also identified, at increasing frequencies, within the An. gambiae s.s. populations from 2012 to 2017, including the SNP Ace1-G280S and its associated duplication. Variants simultaneously identified in the same vector populations raise concerns about the long-term efficacy of new generation bed nets and the recently organophosphate pirimiphos-methyl indoor residual spraying in Burkina Faso.
Conclusion: These findings highlighted the benefit of genomic surveillance of malaria vectors for the detection of new insecticide resistance variants, the monitoring of the existing resistance variants, and also to get insights into the evolutionary processes driving insecticide resistance.
Keywords: An. gambiae; Genomic surveillance; Insecticide resistance; Malaria.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Longitudinal survey of insecticide resistance in a village of central region of Burkina Faso reveals co-occurrence of 1014F, 1014S and 402L mutations in Anopheles coluzzii and Anopheles arabiensis.Malar J. 2024 Aug 20;23(1):250. doi: 10.1186/s12936-024-05069-9. Malar J. 2024. PMID: 39164725 Free PMC article.
-
Species composition and insecticide resistance in malaria vectors in Ellibou, southern Côte d'Ivoire and first finding of Anopheles arabiensis in Côte d'Ivoire.Malar J. 2023 Mar 13;22(1):93. doi: 10.1186/s12936-023-04456-y. Malar J. 2023. PMID: 36915098 Free PMC article.
-
Insecticide resistance status of malaria vectors Anopheles gambiae (s.l.) of southwest Burkina Faso and residual efficacy of indoor residual spraying with microencapsulated pirimiphos-methyl insecticide.Parasit Vectors. 2021 Jan 18;14(1):58. doi: 10.1186/s13071-020-04563-8. Parasit Vectors. 2021. PMID: 33461621 Free PMC article.
-
Dynamics and monitoring of insecticide resistance in malaria vectors across mainland Tanzania from 1997 to 2017: a systematic review.Malar J. 2019 Mar 26;18(1):102. doi: 10.1186/s12936-019-2738-6. Malar J. 2019. PMID: 30914051 Free PMC article.
-
Insecticide resistance in Bemisia tabaci Gennadius (Homoptera: Aleyrodidae) and Anopheles gambiae Giles (Diptera: Culicidae) could compromise the sustainability of malaria vector control strategies in West Africa.Acta Trop. 2013 Oct;128(1):7-17. doi: 10.1016/j.actatropica.2013.06.004. Epub 2013 Jun 17. Acta Trop. 2013. PMID: 23792227 Review.
Cited by
-
A multi-omic meta-analysis reveals novel mechanisms of insecticide resistance in malaria vectors.Commun Biol. 2025 May 23;8(1):790. doi: 10.1038/s42003-025-08221-6. Commun Biol. 2025. PMID: 40410509 Free PMC article.
-
The Anopheles coluzzii range extends into Kenya: Detection, insecticide resistance profiles and population genetic structure in relation to conspecific populations in West and Central Africa.Res Sq [Preprint]. 2024 Feb 15:rs.3.rs-3953608. doi: 10.21203/rs.3.rs-3953608/v1. Res Sq. 2024. Update in: Malar J. 2024 Apr 26;23(1):122. doi: 10.1186/s12936-024-04950-x. PMID: 38410447 Free PMC article. Updated. Preprint.
-
Genomic surveillance reveals geographical heterogeneity and differences in known and novel insecticide resistance mechanisms in Anopheles arabiensis across Kenya.BMC Genomics. 2025 Jul 1;26(1):599. doi: 10.1186/s12864-025-11788-3. BMC Genomics. 2025. PMID: 40597647 Free PMC article.
-
Genomic diversity of the African malaria vector Anopheles funestus.bioRxiv [Preprint]. 2024 Dec 17:2024.12.14.628470. doi: 10.1101/2024.12.14.628470. bioRxiv. 2024. PMID: 39763861 Free PMC article. Preprint.
-
The Anopheles coluzzii range extends into Kenya: detection, insecticide resistance profiles and population genetic structure in relation to conspecific populations in West and Central Africa.Malar J. 2024 Apr 26;23(1):122. doi: 10.1186/s12936-024-04950-x. Malar J. 2024. PMID: 38671462 Free PMC article.
References
-
- WHO. World malaria report 2023. Geneva: World Health Organization; 2023. https://iris.who.int/bitstream/handle/10665/374472/9789240086173-eng.pdf.... Accessed 5 Jan 2024
-
- Epopa PS, Millogo AA, Collins CM, North A, Tripet F, Benedict MQ, et al. The use of sequential mark-release-recapture experiments to estimate population size, survival and dispersal of male mosquitoes of the Anopheles gambiae complex in Bana, a west African humid savannah village. Parasit Vectors. 2017;10:376. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

