Nonrigid registration method for longitudinal chest CT images in COVID-19
- PMID: 39286087
- PMCID: PMC11403531
- DOI: 10.1016/j.heliyon.2024.e37272
Nonrigid registration method for longitudinal chest CT images in COVID-19
Abstract
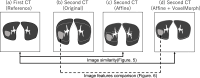
Rationale and objectives: To analyze morphological changes in patients with COVID-19-associated pneumonia over time, a nonrigid registration technique is required that reduces differences in respiratory phase and imaging position and does not excessively deform the lesion region. A nonrigid registration method using deep learning was applied for lung field alignment, and its practicality was verified through quantitative evaluation, such as image similarity of whole lung region and image similarity of lesion region, as well as visual evaluation by a physician.
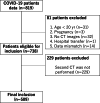
Materials and methods: First, the lung field positions and sizes of the first and second CT images were roughly matched using a classical registration method based on iterative calculations as a preprocessing step. Then, voxel-by-voxel transformation was performed using VoxelMorph, a nonrigid deep learning registration method. As an objective evaluation, the similarity of the images was calculated. To evaluate the invariance of image features in the lesion site, primary statistics and 3D shape features were calculated and statistically analyzed. Furthermore, as a subjective evaluation, the similarity of images and whether nonrigid transformation caused unnatural changes in the shape and size of the lesion region were visually evaluated by a pulmonologist.
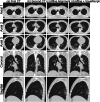
Results: The proposed method was applied to 509 patient data points with high image similarity. The variances in histogram characteristics before and after image deformation were confirmed. Visual evaluation confirmed the agreement between the shape and internal structure of the lung field and the natural deformation of the lesion region.
Conclusion: The developed nonrigid registration method was shown to be effective for quantitative time series analysis of the lungs.
Keywords: Chest computed tomography (CT); Coronavirus disease 2019 (COVID-19); Deep learning; Nonrigid registration.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
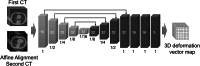



Similar articles
-
Accurate CT∕MR vessel-guided nonrigid registration of largely deformed livers.Med Phys. 2012 May;39(5):2463-77. doi: 10.1118/1.3701779. Med Phys. 2012. PMID: 22559617
-
Position tracking of moving liver lesion based on real-time registration between 2D ultrasound and 3D preoperative images.Med Phys. 2015 Jan;42(1):335-47. doi: 10.1118/1.4903945. Med Phys. 2015. PMID: 25563273
-
Attenuation correction in 4D-PET using a single-phase attenuation map and rigidity-adaptive deformable registration.Med Phys. 2017 Feb;44(2):522-532. doi: 10.1002/mp.12063. Epub 2017 Feb 3. Med Phys. 2017. PMID: 27987223 Free PMC article.
-
CT-PET weighted image fusion for separately scanned whole body rat.Med Phys. 2012 Jan;39(1):533-42. doi: 10.1118/1.3672167. Med Phys. 2012. PMID: 22225323 Free PMC article.
-
Object-constrained meshless deformable algorithm for high speed 3D nonrigid registration between CT and CBCT.Med Phys. 2010 Jan;37(1):197-210. doi: 10.1118/1.3271389. Med Phys. 2010. PMID: 20175482
References
-
- Coronavirus Disease (COVID-19) Situation Reports, (n.d.). https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio... (accessed February 8, 2023).
-
- Suzuki T., Kutsuna S., Nakamura K., Ide S., Moriyama Y., Saito S., Morioka S., Ishikane M., Kinoshita N., Hayakawa K., Ohmagari N. Difficulty of downscaling the precautions for coronavirus disease-19 based on negative throat polymerase chain results in the early phase of infection. J. Infect. Chemother. 2020;26:851–853. doi: 10.1016/J.JIAC.2020.05.002. - DOI - PMC - PubMed
-
- Uwamino Y., Nagata M., Aoki W., Fujimori Y., Nakagawa T., Yokota H., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Shiraki T., Uchida S., Uno S., Kabata H., Ikemura S., Kamata H., Ishii M., Fukunaga K., Kawaoka Y., Hasegawa N., Murata M. Accuracy and stability of saliva as a sample for reverse transcription PCR detection of SARS-CoV-2. J. Clin. Pathol. 2021;74:67–68. doi: 10.1136/JCLINPATH-2020-206972. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

