Multiomic Profiling and Neuroprotective Bioactivity of Salvia Hairy Root-Derived Extracellular Vesicles in a Cellular Model of Parkinson's Disease
- PMID: 39286353
- PMCID: PMC11403015
- DOI: 10.2147/IJN.S479959
Multiomic Profiling and Neuroprotective Bioactivity of Salvia Hairy Root-Derived Extracellular Vesicles in a Cellular Model of Parkinson's Disease
Abstract
Purpose: Extracellular vesicles (EVs) are promising tools for nanomedicine and nanobiotechnology. The purification of mammalian-derived EVs involves intensive processes, and their therapeutic application raises multiple safety and regulatory issues. Plants have the potential to serve as nonconventional sources of therapeutically relevant EVs. In this context, we recently identified hairy roots (HRs) of medicinal plants as a novel biotechnological platform to produce EVs for human health.
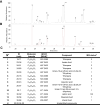
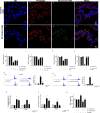
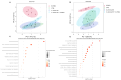
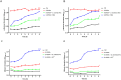
Methods: Herein, we report the purification, omics profiling, and bioactivity of EVs isolated from HRs of the medicinal plants S. sclarea and S. dominica. EVs were isolated from conditioned media of HR cultures using differential ultracentrifugation (dUC) and size exclusion chromatography (SEC). The isolated EVs were characterized by nanoparticle tracking analysis (NTA) and electron microscopy. The proteomic and metabolomic profiles of the EVs were determined using mass spectrometry. Uptake studies and bioactivity assays, including confocal microscopy, MTT, flow cytometry, ROS quantification, and untargeted metabolomics analyses, were conducted in SH-SY5Y cells treated with the neurotoxin 6-hydroxydopamine (6-OHDA) to evaluate the therapeutic potential of EVs in an in vitro model of Parkinson's disease.
Results: S. sclarea HRs released nanosized round-shaped EVs with a distinctive molecular signature. HR EVs from S. sclarea and S. dominica revealed conserved cargo of secondary metabolites, predominantly triterpenoids, which are known for their antioxidant properties. We showed that HR EVs are safe, enter the cells, and strongly inhibit apoptosis in a cellular model of Parkinson's disease. Cellular metabolomics revealed that EVs preserved metabolic homeostasis and mitigated cellular oxidative stress when co-administered with 6-OHDA. Mechanistically, HR EVs inhibited 6-OHDA autoxidation and substantially reduced the accumulation of its oxidative products, which are responsible for 6-OHDA-induced toxicity.
Conclusion: Collectively, our findings provide compelling evidence that EVs isolated from the hairy roots of Salvia species are promising, non-mammalian alternative for the design of novel therapies targeting neurological disorders.
Keywords: Parkinson’s disease; Salvia extracellular vesicles; hairy roots; nanomedicine; neuroprotection; non-mammalian EV source.
© 2024 Vestuto et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest in this work.
Figures



References
-
- Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

