DNA Methylation Carries Signatures of Sublethal Effects Under Thermal Stress in Loggerhead Sea Turtles
- PMID: 39286762
- PMCID: PMC11403127
- DOI: 10.1111/eva.70013
DNA Methylation Carries Signatures of Sublethal Effects Under Thermal Stress in Loggerhead Sea Turtles
Abstract
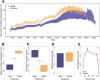
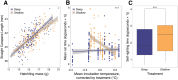
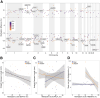
To date, studies of the impacts of climate warming on individuals and populations have mostly focused on mortality and thermal tolerance. In contrast, much less is known about the consequences of sublethal effects, which are more challenging to detect, particularly in wild species with cryptic life histories. This necessitates the development of molecular tools to identify their signatures. In a split-clutch field experiment, we relocated clutches of wild, nesting loggerhead sea turtles (Caretta caretta) to an in situ hatchery. Eggs were then split into two sub-clutches and incubated under shallow or deep conditions, with those in the shallow treatment experiencing significantly higher temperatures in otherwise natural conditions. Although no difference in hatching success was observed between treatments, hatchlings from the shallow, warmer treatment had different length-mass relationships and were weaker at locomotion tests than their siblings incubated in the deep, cooler treatment. To characterise the molecular signatures of these thermal effects, we performed whole genome bisulfite sequencing on blood samples collected upon emergence. We identified 287 differentially methylated sites between hatchlings from different treatments, including on genes with neurodevelopmental, cytoskeletal, and lipid metabolism functions. Taken together, our results show that higher incubation temperatures induce sublethal effects in hatchlings, which are reflected in their DNA methylation status at identified sites. These sites could be used as biomarkers of thermal stress, especially if they are retained across life stages. Overall, this study suggests that global warming reduces hatchling fitness, which has implications for dispersal capacity and ultimately a population's adaptive potential. Conservation efforts for these endangered species and similar climate-threatened taxa will therefore benefit from strategies for monitoring and mitigating exposure to temperatures that induce sublethal effects.
Keywords: DNA methylation; conservation; epigenetics; global warming; sea turtles; sublethal effects; thermal stress.
© 2024 The Author(s). Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
Alice Balard is an Editorial Board member of Evolutionary Applications and a co‐author of this article. To minimise bias, they were excluded from all editorial decision‐making related to the acceptance of this article for publication.
Figures


References
-
- Abdelnour, S. A. , Abd el‐Hack M. E., Khafaga A. F., Arif M., Taha A. E., and Noreldin A. E.. 2019. “Stress Biomarkers and Proteomics Alteration to Thermal Stress in Ruminants: A Review.” Journal of Thermal Biology 79: 120–134. - PubMed
-
- Abella Perez, E. , Marco A., Martins S., and Hawkes L. A.. 2016. “Is This What a Climate Change‐Resilient Population of Marine Turtles Looks Like?” Biological Conservation 193: 124–132.
-
- Akalin, A. , Franke V., Vlahoviček K., Mason C. E., and Schübeler D.. 2015. “Genomation: A Toolkit to Summarize, Annotate and Visualize Genomic Intervals.” Bioinformatics 31: 1127–1129. - PubMed
LinkOut - more resources
Full Text Sources

