G-quadruplexes in an SVA retrotransposon cause aberrant TAF1 gene expression in X-linked dystonia parkinsonism
- PMID: 39287133
- PMCID: PMC12053379
- DOI: 10.1093/nar/gkae797
G-quadruplexes in an SVA retrotransposon cause aberrant TAF1 gene expression in X-linked dystonia parkinsonism
Abstract
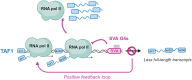
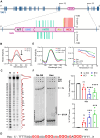
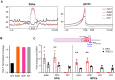
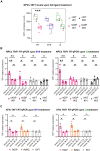
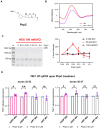
G-quadruplexes (G4s) are non-canonical nucleic acid structures that form in guanine (G)-rich genomic regions. X-linked dystonia parkinsonism (XDP) is an inherited neurodegenerative disease in which a SINE-VNTR-Alu (SVA) retrotransposon, characterised by amplification of a G-rich repeat, is inserted into the coding sequence of TAF1, a key partner of RNA polymerase II. XDP SVA alters TAF1 expression, but the cause of this outcome in XDP remains unknown. To assess whether G4s form in XDP SVA and affect TAF1 expression, we first characterised bioinformatically predicted XDP SVA G4s in vitro. We next showed that highly stable G4s can form and stop polymerase amplification at the SVA region from patient-derived fibroblasts and neural progenitor cells. Using chromatin immunoprecipitazion (ChIP) with an anti-G4 antibody coupled to sequencing or quantitative PCR, we showed that XDP SVA G4s are folded even when embedded in a chromatin context in patient-derived cells. Using the G4 ligands BRACO-19 and quarfloxin and total RNA-sequencing analysis, we showed that stabilisation of the XDP SVA G4s reduces TAF1 transcripts downstream and around the SVA, and increases upstream transcripts, while destabilisation using the G4 unfolder PhpC increases TAF1 transcripts. Our data indicate that G4 formation in the XDP SVA is a major cause of aberrant TAF1 expression, opening the way for the development of strategies to unfold G4s and potentially target the disease.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Lee L.V., Pascasio F.M., Fuentes F.D., Viterbo G.H. Torsion dystonia in Panay, Philippines. Adv. Neurol. 1976; 14:137–151. - PubMed
-
- Arasaratnam C.J., Singh-Bains M.K., Waldvogel H.J., Faull R.L.M. Neuroimaging and neuropathology studies of X-linked dystonia parkinsonism. Neurobiol. Dis. 2021; 148:105186. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

