Altered Cell Clusters and Upregulated Aqp1 in Connexin 50 Knockout Lens Epithelium
- PMID: 39287589
- PMCID: PMC11412383
- DOI: 10.1167/iovs.65.11.27
Altered Cell Clusters and Upregulated Aqp1 in Connexin 50 Knockout Lens Epithelium
Abstract
Purpose: To characterize the heterogeneity and cell clusters of postnatal lens epithelial cells (LECs) and to investigate the downstream targets of connexin 50 (Cx50) in the regulation of lens homeostasis and lens growth. To determine differentially expressed genes (DEGs) in the connexin 50 knockout (Cx50KO) lens epithelial cells that shed light on novel mechanism underlying the cataract and small size of the Cx50KO lenses.
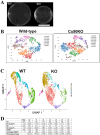
Methods: Single-cell RNA sequencing (scRNA-seq) of lens epithelial cells isolated from one-month-old Cx50KO and wild-type (WT) mice were performed. Differentially expressed genes were identified, and selected DEGs were further studied by quantitative real-time PCR (RT-qPCR) analysis and Western blot analysis.
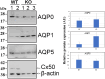
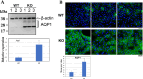
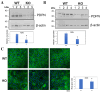
Results: The expression profiles of several thousand genes were identified by scRNA-seq data analysis. In comparison to the WT control, many DEGs were identified in the Cx50KO lens epithelial cells, including growth regulating transcriptional factors and genes encoding water channels. Significantly upregulated aquaporin 1 (Aqp1) gene expression was confirmed by RT-qPCR, and upregulated AQP1 protein expression was confirmed by Western blot analysis and immunostaining both in vivo and in vitro.
Conclusions: Lens epithelial cells exhibit an intrinsic heterogeneity of different cell clusters in regulating lens homeostasis and lens growth. Upregulated Aqp1 in Cx50KO lens epithelial cells suggests that both connexin 50 and AQP1 likely play important roles in regulating water homeostasis in lens epithelial cells.
Conflict of interest statement
Disclosure:
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

