Natural TCRs targeting KRASG12V display fine specificity and sensitivity to human solid tumors
- PMID: 39287991
- PMCID: PMC11529987
- DOI: 10.1172/JCI175790
Natural TCRs targeting KRASG12V display fine specificity and sensitivity to human solid tumors
Abstract
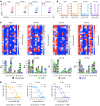
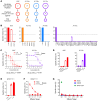
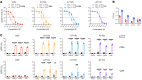
BACKGROUNDNeoantigens derived from KRASMUT have been described, but the fine antigen specificity of T cell responses directed against these epitopes is poorly understood. Here, we explore KRASMUT immunogenicity and the properties of 4 T cell receptors (TCRs) specific for KRASG12V restricted to the HLA-A3 superfamily of class I alleles.METHODSA phase 1 clinical vaccine trial targeting KRASMUT was conducted. TCRs targeting KRASG12V restricted to HLA-A*03:01 or HLA-A*11:01 were isolated from vaccinated patients or healthy individuals. A comprehensive analysis of TCR antigen specificity, affinity, crossreactivity, and CD8 coreceptor dependence was performed. TCR lytic activity was evaluated, and target antigen density was determined by quantitative immunopeptidomics.RESULTSVaccination against KRASMUT resulted in the priming of CD8+ and CD4+ T cell responses. KRASG12V -specific natural (not affinity enhanced) TCRs exhibited exquisite specificity to mutated protein with no discernible reactivity against KRASWT. TCR-recognition motifs were determined and used to identify and exclude crossreactivity to noncognate peptides derived from the human proteome. Both HLA-A*03:01 and HLA-A*11:01-restricted TCR-redirected CD8+ T cells exhibited potent lytic activity against KRASG12V cancers, while only HLA-A*11:01-restricted TCR-T CD4+ T cells exhibited antitumor effector functions consistent with partial coreceptor dependence. All KRASG12V-specific TCRs displayed high sensitivity for antigen as demonstrated by their ability to eliminate tumor cell lines expressing low levels of peptide/HLA (4.4 to 242) complexes per cell.CONCLUSIONThis study identifies KRASG12V-specific TCRs with high therapeutic potential for the development of TCR-T cell therapies.TRIAL REGISTRATIONClinicalTrials.gov NCT03592888.FUNDINGAACR SU2C/Lustgarten Foundation, Parker Institute for Cancer Immunotherapy, and NIH.
Keywords: Antigen presentation; Cancer immunotherapy; Immunology; Oncology; T cell receptor.
Figures



Comment in
- More T cell receptors to the RAScue in cancer? doi: 10.1172/JCI184782
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

