Identification of antibody targets associated with lower HIV viral load and viremic control
- PMID: 39288118
- PMCID: PMC11407625
- DOI: 10.1371/journal.pone.0305976
Identification of antibody targets associated with lower HIV viral load and viremic control
Abstract
Background: High HIV viral loads (VL) are associated with increased morbidity, mortality, and on-going transmission. HIV controllers maintain low VLs in the absence of antiretroviral therapy (ART). We previously used a massively multiplexed antibody profiling assay (VirScan) to compare antibody profiles in HIV controllers and persons living with HIV (PWH) who were virally suppressed on ART. In this report, we used VirScan to evaluate whether antibody reactivity to specific HIV targets and broad reactivity across the HIV genome was associated with VL and controller status 1-2 years after infection.
Methods: Samples were obtained from participants who acquired HIV infection in a community-randomized trial in Africa that evaluated an integrated strategy for HIV prevention (HPTN 071 PopART). Controller status was determined using VL and antiretroviral (ARV) drug data obtained at the seroconversion visit and 1 year later. Viremic controllers had VLs <2,000 copies/mL at both visits; non-controllers had VLs >2,000 copies/mL at both visits. Both groups had no ARV drugs detected at either visit. VirScan testing was performed at the second HIV-positive visit (1-2 years after HIV infection).
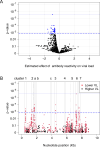
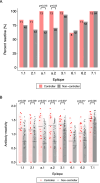
Results: The study cohort included 13 viremic controllers and 64 non-controllers. We identified ten clusters of homologous peptides that had high levels of antibody reactivity (three in gag, three in env, two in integrase, one in protease, and one in vpu). Reactivity to 43 peptides (eight unique epitopes) in six of these clusters was associated with lower VL; reactivity to six of the eight epitopes was associated with HIV controller status. Higher aggregate antibody reactivity across the eight epitopes (more epitopes targeted, higher mean reactivity across all epitopes) and across the HIV genome was also associated with lower VL and controller status.
Conclusions: We identified HIV antibody targets associated with lower VL and HIV controller status 1-2 years after infection. Robust aggregate responses to these targets and broad antibody reactivity across the HIV genome were also associated with lower VL and controller status. These findings provide novel insights into the relationship between humoral immunity and viral containment that could help inform the design of antibody-based approaches for reducing HIV VL.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: H.B.L. is an inventor on an issued patent (US20160320406A) filed by Brigham and Women’s Hospital that covers the use of the VirScan technology, is a founder of Infinity Bio, Portal Bioscience and Alchemab, and is an advisor to TScan Therapeutics.
Figures




References
-
- Fauci AS, Desrosiers RC. Pathogenesis of HIV and SIV. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY)1997. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

