A naturally occurring 22-amino acid fragment of human hemoglobin A inhibits autophagy and HIV-1
- PMID: 39289189
- PMCID: PMC11408460
- DOI: 10.1007/s00018-024-05447-1
A naturally occurring 22-amino acid fragment of human hemoglobin A inhibits autophagy and HIV-1
Abstract
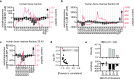
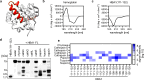
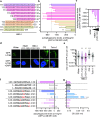
Autophagy is an evolutionarily ancient catabolic pathway and has recently emerged as an integral part of the innate immune system. While the core machinery of autophagy is well defined, the physiological regulation of autophagy is less understood. Here, we identify a C-terminal fragment of human hemoglobin A (HBA1, amino acids 111-132) in human bone marrow as a fast-acting non-inflammatory inhibitor of autophagy initiation. It is proteolytically released from full-length HBA1 by cathepsin E, trypsin or pepsin. Biochemical characterization revealed that HBA1(111-132) has an in vitro stability of 52 min in human plasma and adopts a flexible monomeric conformation in solution. Structure-activity relationship studies revealed that the C-terminal 13 amino acids of HBA1(120-132) are sufficient to inhibit autophagy, two charged amino acids (D127, K128) mediate solubility, and two serines (S125, S132) are required for function. Successful viruses like human immunodeficiency virus 1 (HIV-1) evolved strategies to subvert autophagy for virion production. Our results show that HBA1(120-132) reduced virus yields of lab-adapted and primary HIV-1. Summarizing, our data identifies naturally occurring HBA1(111-132) as a physiological, non-inflammatory antagonist of autophagy. Optimized derivatives of HBA1(111-132) may offer perspectives to restrict autophagy-dependent viruses.
Keywords: Autophagy; HIV; Hemoglobin; Innate immunity.
© 2024. The Author(s).
Conflict of interest statement
DRT collaborated with Novartis Pharma AG (Basel, Switzerland), Probiodrug (Halle (Saale), Germany), GE Healthcare (Amersham, UK), and Janssen Pharmaceutical Companies (Beerse, Belgium). All other authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

