Basal-to-inflammatory transition and tumor resistance via crosstalk with a pro-inflammatory stromal niche
- PMID: 39289380
- PMCID: PMC11408617
- DOI: 10.1038/s41467-024-52394-3
Basal-to-inflammatory transition and tumor resistance via crosstalk with a pro-inflammatory stromal niche
Abstract
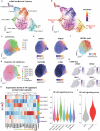
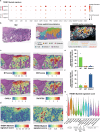
Cancer-associated inflammation is a double-edged sword possessing both pro- and anti-tumor properties through ill-defined tumor-immune dynamics. While we previously identified a carcinoma tumor-intrinsic resistance pathway, basal-to-squamous cell carcinoma transition, here, employing a multipronged single-cell and spatial-omics approach, we identify an inflammation and therapy-enriched tumor state we term basal-to-inflammatory transition. Basal-to-inflammatory transition signature correlates with poor overall patient survival in many epithelial tumors. Basal-to-squamous cell carcinoma transition and basal-to-inflammatory transition occur in adjacent but distinct regions of a single tumor: basal-to-squamous cell carcinoma transition arises within the core tumor nodule, while basal-to-inflammatory transition emerges from a specialized inflammatory environment defined by a tumor-associated TREM1 myeloid signature. TREM1 myeloid-derived cytokines IL1 and OSM induce basal-to-inflammatory transition in vitro and in vivo through NF-κB, lowering sensitivity of patient basal cell carcinoma explant tumors to Smoothened inhibitor treatment. This work deepens our knowledge of the heterogeneous local tumor microenvironment and nominates basal-to-inflammatory transition as a drug-resistant but targetable tumor state driven by a specialized inflammatory microenvironment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Miller, K. D. et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J. Clin.72, 409–436 (2022). - PubMed
-
- Mcmillin, D. W., Negri, J. M. & Mitsiades, C. S. The role of tumour-stromal interactions in modifying drug response: Challenges and opportunities. Nat. Rev. Drug Discov.12, 217–228 (2013). - PubMed
-
- Hanahan, D. & Coussens, L. M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell21, 309–322 (2012). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- S10 OD025212/OD/NIH HHS/United States
- R37 AR054780/AR/NIAMS NIH HHS/United States
- U54 CA274511/CA/NCI NIH HHS/United States
- S10 OD021763/OD/NIH HHS/United States
- U54 CA209971/CA/NCI NIH HHS/United States
- S10 OD010580/OD/NIH HHS/United States
- S10 RR027431/RR/NCRR NIH HHS/United States
- T32 AR007422/AR/NIAMS NIH HHS/United States
- P30 CA062203/CA/NCI NIH HHS/United States
- F32 CA254434/CA/NCI NIH HHS/United States
- 2R37-ARO54780/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- S10 RR025518/RR/NCRR NIH HHS/United States
- RO1 ARO46786/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

