The European Reference Genome Atlas: piloting a decentralised approach to equitable biodiversity genomics
- PMID: 39289538
- PMCID: PMC11408602
- DOI: 10.1038/s44185-024-00054-6
The European Reference Genome Atlas: piloting a decentralised approach to equitable biodiversity genomics
Erratum in
-
Author Correction: The European Reference Genome Atlas: piloting a decentralised approach to equitable biodiversity genomics.NPJ Biodivers. 2024 Oct 15;3(1):31. doi: 10.1038/s44185-024-00065-3. NPJ Biodivers. 2024. PMID: 39407030 Free PMC article. No abstract available.
Abstract
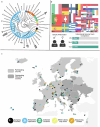
A genomic database of all Earth's eukaryotic species could contribute to many scientific discoveries; however, only a tiny fraction of species have genomic information available. In 2018, scientists across the world united under the Earth BioGenome Project (EBP), aiming to produce a database of high-quality reference genomes containing all ~1.5 million recognized eukaryotic species. As the European node of the EBP, the European Reference Genome Atlas (ERGA) sought to implement a new decentralised, equitable and inclusive model for producing reference genomes. For this, ERGA launched a Pilot Project establishing the first distributed reference genome production infrastructure and testing it on 98 eukaryotic species from 33 European countries. Here we outline the infrastructure and explore its effectiveness for scaling high-quality reference genome production, whilst considering equity and inclusion. The outcomes and lessons learned provide a solid foundation for ERGA while offering key learnings to other transnational, national genomic resource projects and the EBP.
© 2024. The Author(s).
Conflict of interest statement
Jean-François Flot, Rosa Fernández, Javier Del Campo, Josefa Gonzáles, Olga Vinnere Pettersson, Robert M Watherhouse, Patrick Wincker and Sylke Winkler are recommenders for PCI Genomics. The authors declare they have no further conflict of interest relating to the content of this article.
Figures



References
-
- UNEP. Facts about the nature crisis. UNEP—UN Environment Programmehttps://www.unep.org/facts-about-nature-crisis (2022).
-
- Zhang, Y., Wang, Z., Lu, Y. & Zuo, L. Editorial: biodiversity, ecosystem functions and services: Interrelationship with environmental and human health. Front. Ecol. Evol. 10, 10.3389/fevo.2022.1086408 (2022).
-
- IUCN. The IUCN Red List of Threatened Species Version 2022-2. The IUCN Red List of Threatened Specieshttps://www.iucnredlist.org.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

