Vascular heterogeneity of tight junction Claudins guides organotropic metastasis
- PMID: 39289595
- PMCID: PMC11987010
- DOI: 10.1038/s43018-024-00813-1
Vascular heterogeneity of tight junction Claudins guides organotropic metastasis
Abstract
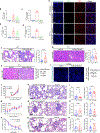
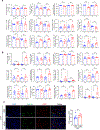
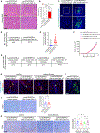
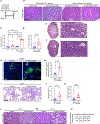
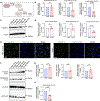
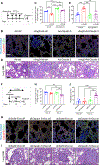
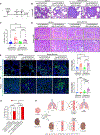
Carcinomas are associated with metastasis to specific organs while sparing others. Breast cancer presents with lung metastasis but rarely kidney metastasis. Using this difference as an example, we queried the mechanism(s) behind the proclivity for organ-specific metastasis. We used spontaneous and implant models of metastatic mammary carcinoma coupled with inflammatory tissue fibrosis, single-cell sequencing analyses and functional studies to unravel the causal determinants of organ-specific metastasis. Here we show that lung metastasis is facilitated by angiopoietin 2 (Ang2)-mediated suppression of lung-specific endothelial tight junction protein Claudin 5, which is augmented by the inflammatory fibrotic microenvironment and prevented by anti-Ang2 blocking antibodies, while kidney metastasis is prevented by non-Ang2-responsive Claudins 2 and 10. Suppression of Claudins 2 and 10 was sufficient to induce the emergence of kidney metastasis. This study illustrates the influence of organ-specific vascular heterogeneity in determining organotropic metastasis, independent of cancer cell-intrinsic mechanisms.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests Statement
The authors declare no conflict of interest relevant to this manuscript.
Figures











References
-
- Paget S The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev 8, 98–101 (1989). - PubMed
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- RP200612/Cancer Prevention and Research Institute of Texas (Cancer Prevention Research Institute of Texas)
- R01 CA252729/CA/NCI NIH HHS/United States
- R01CA252729/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P30CA016672/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

