Real-world experience with circulating tumor DNA in cerebrospinal fluid from patients with central nervous system tumors
- PMID: 39289779
- PMCID: PMC11406943
- DOI: 10.1186/s40478-024-01846-4
Real-world experience with circulating tumor DNA in cerebrospinal fluid from patients with central nervous system tumors
Abstract
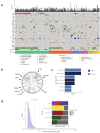
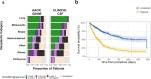
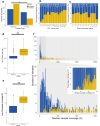
The characterization of genetic alterations in tumor samples has become standard practice for many human cancers to achieve more precise disease classification and guide the selection of targeted therapies. Cerebrospinal fluid (CSF) can serve as a source of tumor DNA in patients with central nervous system (CNS) cancer. We performed comprehensive profiling of CSF circulating tumor DNA (ctDNA) in 711 patients using an FDA-authorized platform (MSK-IMPACT™) in a hospital laboratory. We identified genetic alterations in 489/922 (53.0%) CSF samples with clinically documented CNS tumors. None of 85 CSF samples from patients without CNS tumors had detectable ctDNA. The distribution of clinically actionable somatic alterations was consistent with tumor-type specific alterations across the AACR GENIE cohort. Repeated CSF ctDNA examinations from the same patients identified clonal evolution and emergence of resistance mechanisms. ctDNA detection was associated with shortened overall survival following CSF collection. Next-generation sequencing of CSF, collected through a minimally invasive lumbar puncture in a routine hospital setting, provides clinically actionable cancer genotype information in a large fraction of patients with CNS tumors.
© 2024. The Author(s).
Conflict of interest statement
IKM reports receiving personal fees (advisory board services) from Agios, Black Diamond Therapeutics, Debiopharm Group, Erasca, Novartis, Prelude Therapeutics, Roche Therapeutics, Servier Pharmaceuticals, Voyager. IKM reports receiving grants from General Electric and Puma Biotechnology. MEA reports consulting for Janssen Global Services, Bristol-Myers Squibb, AstraZeneca, Roche, Biocartis, Sanofi. MEA also reports personal fees (advisory roles) for Invivoscribe, Physician Educational Resources (PER), Peerview Institute for medical education, clinical care options, and RMEI medical education. RAH is an employee of Foundation Medicine, Inc., a wholly owned subsidiary of Roche holdings, Inc. and Roche Finance Ltd., and has equity interest in an affiliate of these Roche holdings.
Figures

References
-
- Aizer AA, Lamba N, Ahluwalia MS, Aldape K, Boire A, Brastianos PK, Brown PD, Camidge DR, Chiang VL, Davies MA et al (2022) Brain metastases: a society for neuro-oncology (SNO) consensus review on current management and future directions. Neuro Oncol 24:1613–1646. 10.1093/neuonc/noac118 - PMC - PubMed
-
- Bale TA, Yang S-R, Solomon JP, Nafa K, Middha S, Casanova J, Sadowska J, Skakodub A, Ahmad H, Helena AY (2021) Clinical experience of cerebrospinal fluid-based liquid biopsy demonstrates superiority of cell-free DNA over cell pellet genomic DNA for molecular profiling. J Mol Diagn 23:742–752 - PMC - PubMed
-
- Bale TA, Yang SR, Solomon JP, Nafa K, Middha S, Casanova J, Sadowska J, Skakodub A, Ahmad H, Yu HA et al (2021) Clinical experience of cerebrospinal fluid-based liquid biopsy demonstrates superiority of cell-free DNA over cell pellet genomic DNA for molecular profiling. J. Mol. Diagn. JMD 23:742–752. 10.1016/j.jmoldx.2021.03.001 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

