Transmission dynamics of ESBL/AmpC and carbapenemase-producing Enterobacterales between companion animals and humans
- PMID: 39290515
- PMCID: PMC11405340
- DOI: 10.3389/fmicb.2024.1432240
Transmission dynamics of ESBL/AmpC and carbapenemase-producing Enterobacterales between companion animals and humans
Abstract
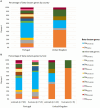
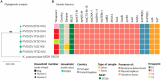
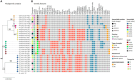
Antimicrobial resistance mediated by extended-spectrum beta-lactamase (ESBL)- and plasmid-mediated cephalosporinase (AmpC)-producing Enterobacterales, as well as carbapenemase-producing Enterobacterales have globally increased among companion animals, posing a potential health risk to humans in contact with them. This prospective longitudinal study investigates the transfer of ESBL/AmpC- and carbapenemase-producing Enterobacterales between companion animals and their cohabitant humans in Portugal (PT) and the United Kingdom (UK) during animal infection. Fecal samples and nasal swabs collected from dogs and cats with urinary tract infection (UTI) or skin and soft tissue infection (SSTI), and their cohabitant humans were screened for resistant strains. Relatedness between animal and human strains was established by whole-genome sequencing (WGS). ESBL/AmpC-producing Enterobacterales were detected in companion animals (PT = 55.8%; UK = 36.4%) and humans (PT = 35.9%; UK = 12.5%). Carbapenemase-producing Enterobacterales carriage was observed in one dog from Portugal (2.6%) and another dog from the UK (4.5%). Transmission of index clinical ESBL-producing Escherichia coli and Klebsiella pneumoniae strains to cohabitant humans was observed in three Portuguese households (6.9%, n = 43), with repeated isolation of the index strains on fecal samples from the animals and their cohabiting humans. In addition, longitudinal sharing of E. coli strains carried by companion animals and their owners was observed in other two Portuguese households and two households from the UK. Furthermore, a multidrug-resistant ACT-24-producing Enterobacter hormaechei subsp. hoffmannii strains were also shared within another Portuguese household. These results highlight the importance of the household as an epidemiological unit in the efforts to mitigate the spread of antimicrobial resistance, further emphasizing the need for antimicrobial surveillance in this context, capable of producing data that can inform and evaluate public health actions.
Keywords: CMY-2; CTX-M-15 ESBL; CTX-M-27; Enterobacter hormaechei subsp. hoffmannii; ExPEC pathotypes; Klebsiella pneumoniae; animal–human sharing; one health.
Copyright © 2024 Menezes, Frosini, Weese, Perreten, Schwarz, Amaral, Loeffler and Pomba.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Abbas G., Khan I., Mohsin M., Sajjad-ur-Rahman S. U. R., Younas T., Ali S. (2019). High rates of CTX-M group-1 extended-spectrum β-lactamases producing Escherichia coli from pets and their owners in Faisalabad. Pakistan. Infect. Drug Resist. 12, 571–578. doi: 10.2147/IDR.S189884, PMID: - DOI - PMC - PubMed
-
- Andrews S. (2010) A quality control tool for high throughput sequence data. Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed: 10 January 2021).
-
- Baede V. O., Broens E. M., Spaninks M. P., Timmerman A. J., Graveland H., Wagenaar J. A., et al. (2017). Raw pet food as a risk factor for shedding of extended-spectrum beta-lactamase-producing Enterobacteriaceae in household cats. PLoS One 12:e0187239. doi: 10.1371/journal.pone.0187239, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

