Different Biomechanical Cell Behaviors in an Epithelium Drive Collective Epithelial Cell Extrusion
- PMID: 39291385
- PMCID: PMC11558136
- DOI: 10.1002/advs.202401573
Different Biomechanical Cell Behaviors in an Epithelium Drive Collective Epithelial Cell Extrusion
Abstract
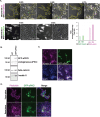
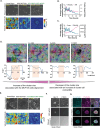
In vertebrates, many organs, such as the kidney and the mammary gland form ductal structures based on the folding of epithelial sheets. The development of these organs relies on coordinated sorting of different cell lineages in both time and space, through mechanisms that remain largely unclear. Tissues are composed of several cell types with distinct biomechanical properties, particularly at cell-cell and cell-substrate boundaries. One hypothesis is that adjacent epithelial layers work in a coordinated manner to shape the tissue. Using in vitro experiments on model epithelial cells, differential expression of atypical Protein Kinase C iota (aPKCi), a key junctional polarity protein, is shown to reinforce cell epithelialization and trigger sorting by tuning cell mechanical properties at the tissue level. In a broader perspective, it is shown that in a heterogeneous epithelial monolayer, in which cell sorting occurs, forces arising from epithelial cell growth under confinement by surrounding cells with different biomechanical properties are sufficient to promote collective cell extrusion and generate emerging 3D organization related to spheroids and buds. Overall, this research sheds light on the role of aPKCi and the biomechanical interplay between distinct epithelial cell lineages in shaping tissue organization, providing insights into the understanding of tissue and organ development.
Keywords: Proc Natl Acad Sci USA; cell sorting; collective cell extrusion; dewetting; mechanobiology.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
