Hybrid modeling for in silico optimization of a dynamic perfusion cell culture process
- PMID: 39291457
- PMCID: PMC11831417
- DOI: 10.1002/btpr.3503
Hybrid modeling for in silico optimization of a dynamic perfusion cell culture process
Abstract
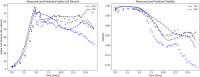
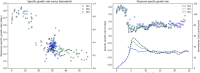
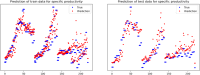
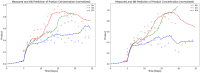
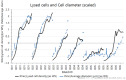
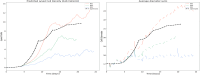
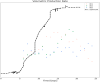
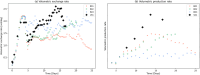
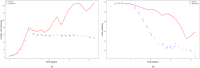
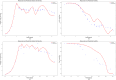
The bio-pharmaceutical industry heavily relies on mammalian cells for the production of bio-therapeutic proteins. The complexity of implementing and high cost-of-goods of these processes are currently limiting more widespread patient access. This is driving efforts to enhance cell culture productivity and cost reduction. Upstream process intensification (PI), using perfusion approaches in the seed train and/or the main bioreactor, has shown substantial promise to enhance productivity. However, developing optimal process conditions for perfusion-based processes remain challenging due to resource and time constraints. Model-based optimization offers a solution by systematically screening process parameters like temperature, pH, and culture media to find the optimum conditions in silico. To our knowledge, this is the first experimentally validated model to explain the perfusion dynamics under different operating conditions and scales for process optimization. The hybrid model accurately describes Chinese hamster ovary (CHO) cell culture growth dynamics and a neural network model explains the production of mAb, allowing for optimization of media exchange rates. Results from six perfusion runs in Ambr® 250 demonstrated high accuracy, confirming the model's utility. Further, the implementation of dynamic media exchange rate schedule determined through model-based optimization resulted in 50% increase in volumetric productivity. Additionally, two 5 L-scale experiments validated the model's reliable extrapolation capabilities to large bioreactors. This approach could reduce the number of wet lab experiments needed for culture process optimization, offering a promising avenue for improving productivity, cost-of-goods in bio-pharmaceutical manufacturing, in turn improving patient access to pivotal medicine.
Keywords: CHO cells; digital twin; hybrid models; modeling; optimization; perfusion.
© 2024 The Author(s). Biotechnology Progress published by Wiley Periodicals LLC on behalf of American Institute of Chemical Engineers.
Conflict of interest statement
Piyush Agarwal (at the time of manuscript submission), Chris McCready, and Gerben Zijlstra were Sartorius employees. Maarten Pennings, Jeroen van de Laar, and Jake Chng Ng were BiosanaPharma employees and Say Kong Ng was BTI employee. The authors declare no conflict of interest.
Figures
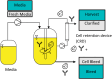

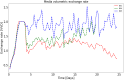
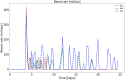
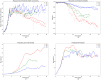











References
-
- Yeo HC, Park S‐Y, Tan T, Ng SK, Lakshmanan M, Lee D‐Y. Combined multivariate statistical and flux balance analyses uncover media bottlenecks to the growth and productivity of Chinese hamster ovary cell cultures. Biotechnol Bioeng. 2022;119(7):1740‐1754. - PubMed
-
- MacDonald MA, Nöbel M, Roche Recinos D, et al. Perfusion culture of Chinese hamster ovary cells for bioprocessing applications. Crit Rev Biotechnol. 2022;42(7):1099‐1115. - PubMed
-
- Müller D, Klein L, Lemke J, et al. Process intensification in the biopharma industry: improving efficiency of protein manufacturing processes from development to production scale using synergistic approaches. Chem Eng Process‐Process Intensification. 2022;171:108727.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

