Amylase-associated genetic pattern in Xanthomonas euvesicatoria on pepper
- PMID: 39291986
- PMCID: PMC11497833
- DOI: 10.1128/aem.01313-24
Amylase-associated genetic pattern in Xanthomonas euvesicatoria on pepper
Abstract
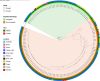
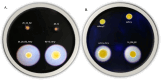
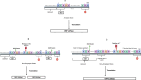
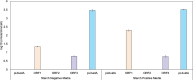
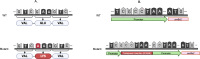
Bacterial leaf spot of pepper (BSP), primarily caused by Xanthomonas euvesicatoria (Xe), poses a significant challenge to pepper production worldwide. Despite its impact, the genetic diversity of this pathogen remains underexplored, which limits our understanding of its population structure. To bridge this knowledge gap, we conducted a comprehensive analysis using 103 Xe strains isolated from pepper in southwest Florida to characterize genomic and type III effector (T3E) variation in this population. Phylogenetic analysis of core genomes revealed a major distinct genetic lineage associated with amylolytic activity. This amylolytic lineage was represented in Xe strains globally. Molecular clock analysis dated the emergence of amylolytic strains in Xe to around 1972. Notably, non-amylolytic strains possessed a single base pair frameshift deletion in the ⍺-amylase gene yet retained a conserved C-terminus. GUS assay revealed the expression of two open reading frames in non-amylolytic strains, one at the N-terminus and another that starts 136 base pairs upstream of the ⍺-amylase gene. Analysis of T3Es in the Florida Xe population identified variation in 12 effectors, including two classes of mutations in avrBs2 that prevent AvrBs2 from triggering a hypersensitive response in Bs2-resistant pepper plants. Knowledge of T3E variation could be used for effector-targeted disease management. This study revealed previously undescribed population structure in this economically important pathogen.IMPORTANCEBacterial leaf spot (BSP), a significant threat to pepper production globally, is primarily caused by Xanthomonas euvesicatoria (Xe). Limited genomic data has hindered detailed studies on its population diversity. This study analyzed the whole-genome sequences of 103 Xe strains from peppers in southwest Florida, along with additional global strains, to explore the pathogen's diversity. The study revealed two major distinct genetic groups based on their amylolytic activity, the ability to break down starch, with non-amylolytic strains having a mutation in the ⍺-amylase gene. Additionally, two classes of mutations in the avrBs2 gene were found, leading to susceptibility in pepper plants with the Bs2 resistance gene, a commercially available resistance gene for BSP. These findings highlight the need to forecast the emergence of such strains, identify genetic factors for innovative disease management, and understand how this pathogen evolves and spreads.
Keywords: Pepper; Xanthomonas euvesicatoria; amylase; frameshift; population genomics; type III effectors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Cesbron S, Briand M, Essakhi S, Gironde S, Boureau T, Manceau C, Fischer-Le Saux M, Jacques MA. 2015. Comparative Genomics of pathogenic and nonpathogenic strains of Xanthomonas arboricola unveil molecular and evolutionary events linked to pathoadaptation. Front Plant Sci 6:1126. doi: 10.3389/fpls.2015.01126 - DOI - PMC - PubMed
-
- Mhedbi-Hajri N, Hajri A, Boureau T, Darrasse A, Durand K, Brin C, Fischer-Le Saux M, Manceau C, Poussier S, Pruvost O, Lemaire C, Jacques M-A. 2013. Evolutionary history of the plant pathogenic bacterium Xanthomonas axonopodis. PLoS ONE 8:e58474. doi: 10.1371/journal.pone.0058474 - DOI - PMC - PubMed
-
- Richard D, Ravigné V, Rieux A, Facon B, Boyer C, Boyer K, Grygiel P, Javegny S, Terville M, Canteros BI, Robène I, Vernière C, Chabirand A, Pruvost O, Lefeuvre P. 2017. Adaptation of genetically monomorphic bacteria: evolution of copper resistance through multiple horizontal gene transfers of complex and versatile mobile genetic elements. Mol Ecol 26:2131–2149. doi: 10.1111/mec.14007 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

