MultiGreen: A multiplexing architecture for GreenGate cloning
- PMID: 39292669
- PMCID: PMC11410190
- DOI: 10.1371/journal.pone.0306008
MultiGreen: A multiplexing architecture for GreenGate cloning
Abstract
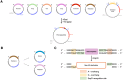
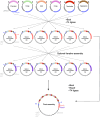
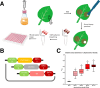
Genetic modification of plants fundamentally relies upon customized vector designs. The ever-increasing complexity of transgenic constructs has led to increased adoption of modular cloning systems for their ease of use, cost effectiveness, and rapid prototyping. GreenGate is a modular cloning system catered specifically to designing bespoke, single transcriptional unit vectors for plant transformation-which is also its greatest flaw. MultiGreen seeks to address GreenGate's limitations while maintaining the syntax of the original GreenGate kit. The primary limitations MultiGreen addresses are 1) multiplexing in series, 2) multiplexing in parallel, and 3) repeated cycling of transcriptional unit assembly through binary intermediates. MultiGreen efficiently concatenates bespoke transcriptional units using an additional suite of level 1acceptor vectors which serve as an assembly point for individual transcriptional units prior to final, level 2, condensation of multiple transcriptional units. Assembly with MultiGreen level 1 vectors scales at a maximal rate of 2*⌈log6n⌉+3 days per assembly, where n represents the number of transcriptional units. Further, MultiGreen level 1 acceptor vectors are binary vectors and can be used directly for plant transformation to further maximize prototyping speed. MultiGreen is a 1:1 expansion of the original GreenGate architecture's grammar and has been demonstrated to efficiently assemble plasmids with multiple transcriptional units. MultiGreen has been validated by using a truncated violacein operon from Chromobacterium violaceum in bacteria and by deconstructing the RUBY reporter for in planta functional validation. MultiGreen currently supports many of our in-house multi transcriptional unit assemblies and will be a valuable strategy for more complex cloning projects.
Copyright: © 2024 Pennetti et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Bolivar F, Backman K. [16] Plasmids of Escherichia coli as cloning vectors. Methods in enzymology. 68: Elsevier; 1979. p. 245–67. - PubMed
-
- Rogers SG, Klee H, Horsch R, Fraley R. [15] Improved vectors for plant transformation: Expression cassette vectors and new selectable markers. Methods in Enzymology. 153: Elsevier; 1987. p. 253–77.
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

