Isolation and characterization of IgG3 glycan-targeting antibodies with exceptional cross-reactivity for diverse viral families
- PMID: 39292703
- PMCID: PMC11410209
- DOI: 10.1371/journal.ppat.1012499
Isolation and characterization of IgG3 glycan-targeting antibodies with exceptional cross-reactivity for diverse viral families
Abstract
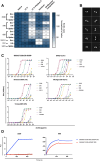
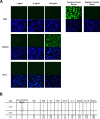
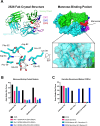
Broadly reactive antibodies that target sequence-diverse antigens are of interest for vaccine design and monoclonal antibody therapeutic development because they can protect against multiple strains of a virus and provide a barrier to evolution of escape mutants. Using LIBRA-seq (linking B cell receptor to antigen specificity through sequencing) data for the B cell repertoire of an individual chronically infected with human immunodeficiency virus type 1 (HIV-1), we identified a lineage of IgG3 antibodies predicted to bind to HIV-1 Envelope (Env) and influenza A Hemagglutinin (HA). Two lineage members, antibodies 2526 and 546, were confirmed to bind to a large panel of diverse antigens, including several strains of HIV-1 Env, influenza HA, coronavirus (CoV) spike, hepatitis C virus (HCV) E protein, Nipah virus (NiV) F protein, and Langya virus (LayV) F protein. We found that both antibodies bind to complex glycans on the antigenic surfaces. Antibody 2526 targets the stem region of influenza HA and the N-terminal domain (NTD) region of SARS-CoV-2 spike. A crystal structure of 2526 Fab bound to mannose revealed the presence of a glycan-binding pocket on the light chain. Antibody 2526 cross-reacted with antigens from multiple pathogens and displayed no signs of autoreactivity. These features distinguish antibody 2526 from previously described glycan-reactive antibodies. Further study of this antibody class may aid in the selection and engineering of broadly reactive antibody therapeutics and can inform the development of effective vaccines with exceptional breadth of pathogen coverage.
Copyright: © 2024 Vukovich et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
M.J.V., A.R.S., and I.S.G. are listed as inventors on patents filed describing the antibodies discovered here. I.S.G. is listed as an inventor on the patent applications for the LIBRA-seq technology. I.S.G. is a co-founder of AbSeek Bio. I.S.G. has served as a consultant for Sanofi. The Georgiev laboratory at VUMC has received unrelated funding from Merck and Takeda Pharmaceuticals. D.J.S has served as a consultant for AstraZeneca AB.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

