Targeting NAT10 inhibits osteosarcoma progression via ATF4/ASNS-mediated asparagine biosynthesis
- PMID: 39293390
- PMCID: PMC11525028
- DOI: 10.1016/j.xcrm.2024.101728
Targeting NAT10 inhibits osteosarcoma progression via ATF4/ASNS-mediated asparagine biosynthesis
Abstract
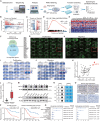
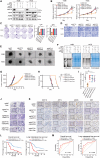
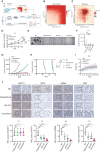
Despite advances in treatment, the prognosis of patients with osteosarcoma remains unsatisfactory, and searching for potential targets is imperative. Here, we identify N4-acetylcytidine (ac4C) acetyltransferase 10 (NAT10) as a candidate therapeutic target in osteosarcoma through functional screening. NAT10 overexpression is correlated with a poor prognosis, and NAT10 knockout inhibits osteosarcoma progression. Mechanistically, NAT10 enhances mRNA stability of activating transcription factor 4 (ATF4) through ac4C modification. ATF4 induces the transcription of asparagine synthetase (ASNS), which catalyzes asparagine (Asn) biosynthesis, facilitating osteosarcoma progression. Utilizing virtual screening, we identify paliperidone and AG-401 as potential NAT10 inhibitors, and both inhibitors are found to bind to NAT10 proteins. Inhibiting NAT10 suppresses osteosarcoma progression in vivo. Combined treatment using paliperidone and AG-401 produces synergistic inhibition for osteosarcoma in patient-derived xenograft (PDX) models. Our findings demonstrate that NAT10 facilitates osteosarcoma progression through the ATF4/ASNS/Asn axis, and pharmacological inhibition of NAT10 may be a feasible therapeutic approach for osteosarcoma.
Keywords: ASNS; ATF4; N4-acetylcytidine; NAT10; asparagine; osteosarcoma.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

