Radiomics in breast cancer: Current advances and future directions
- PMID: 39293402
- PMCID: PMC11528234
- DOI: 10.1016/j.xcrm.2024.101719
Radiomics in breast cancer: Current advances and future directions
Abstract
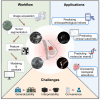
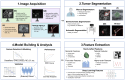
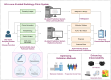
Breast cancer is a common disease that causes great health concerns to women worldwide. During the diagnosis and treatment of breast cancer, medical imaging plays an essential role, but its interpretation relies on radiologists or clinical doctors. Radiomics can extract high-throughput quantitative imaging features from images of various modalities via traditional machine learning or deep learning methods following a series of standard processes. Hopefully, radiomic models may aid various processes in clinical practice. In this review, we summarize the current utilization of radiomics for predicting clinicopathological indices and clinical outcomes. We also focus on radio-multi-omics studies that bridge the gap between phenotypic and microscopic scale information. Acknowledging the deficiencies that currently hinder the clinical adoption of radiomic models, we discuss the underlying causes of this situation and propose future directions for advancing radiomics in breast cancer research.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
References
-
- Lambin P., Rios-Velazquez E., Leijenaar R., Carvalho S., van Stiphout R.G.P.M., Granton P., Zegers C.M.L., Gillies R., Boellard R., Dekker A., Aerts H.J.W.L. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

