Congenital microcoria deletion in mouse links Sox21 dysregulation to disease and suggests a role for TGFB2 in glaucoma and myopia
- PMID: 39293448
- PMCID: PMC11480854
- DOI: 10.1016/j.ajhg.2024.08.019
Congenital microcoria deletion in mouse links Sox21 dysregulation to disease and suggests a role for TGFB2 in glaucoma and myopia
Abstract
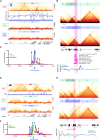
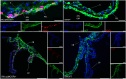
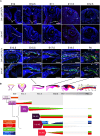
Congenital microcoria (MCOR) is a rare hereditary developmental defect of the iris dilator muscle frequently associated with high axial myopia and high intraocular pressure (IOP) glaucoma. The condition is caused by submicroscopic rearrangements of chromosome 13q32.1. However, the mechanisms underlying the failure of iris development and the origin of associated features remain elusive. Here, we present a 3D architecture model of the 13q32.1 region, demonstrating that MCOR-related deletions consistently disrupt the boundary between two topologically associating domains (TADs). Deleting the critical MCOR-causing region in mice reveals ectopic Sox21 expression precisely aligning with Dct, each located in one of the two neighbor TADs. This observation is consistent with the TADs' boundary alteration and adoption of Dct regulatory elements by the Sox21 promoter. Additionally, we identify Tgfb2 as a target gene of SOX21 and show TGFΒ2 accumulation in the aqueous humor of an MCOR-affected subject. Accumulation of TGFB2 is recognized for its role in glaucoma and potential impact on axial myopia. Our results highlight the importance of SOX21-TGFB2 signaling in iris development and control of eye growth and IOP. Insights from MCOR studies may provide therapeutic avenues for this condition but also for glaucoma and high myopia conditions, affecting millions of people.
Keywords: DCT enhancer adoption; SOX21-TGFB2 signalling glaucoma and myopia; Sox21 ectopic expression; congenital microcoria; developmental genetics; genetic eye disorders; genome architecture; mouse model; topologically-associated domain; translational medicine.
Copyright © 2024 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The SOX21-TGFB2 pathway in iris development, axial myopia, and GLC has been officially patented under the title “Methods and pharmaceutical compositions for treating ocular diseases” (WO/2021/245224). The inventors of this patent are J.-M.R., L.F.T., B.N., C.A., and J.K.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

