Transient hypoxia drives soil microbial community dynamics and biogeochemistry during human decomposition
- PMID: 39293810
- PMCID: PMC11879408
- DOI: 10.1093/femsec/fiae119
Transient hypoxia drives soil microbial community dynamics and biogeochemistry during human decomposition
Abstract
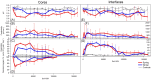
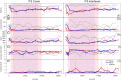
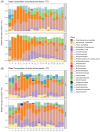
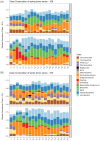
Human decomposition in terrestrial ecosystems is a dynamic process creating localized hot spots of soil microbial activity. Longer-term (beyond a few months) impacts on decomposer microbial communities are poorly characterized and do not typically connect microbial communities to biogeochemistry, limiting our understanding of decomposer communities and their functions. We performed separate year-long human decomposition trials, one starting in spring, another in winter, integrating bacterial and fungal community structure and abundances with soil physicochemistry and biogeochemistry to identify key drivers of microbial community change. In both trials, soil acidification, elevated microbial respiration, and reduced soil oxygen concentrations occurred. Changes in soil oxygen concentrations were the primary driver of microbial succession and nitrogen transformation patterns, while fungal community diversity and abundance was related to soil pH. Relative abundance of facultative anaerobic taxa (Firmicutes and Saccharomycetes) increased during the period of reduced soil oxygen. The magnitude and timing of the decomposition responses were amplified during the spring trial relative to the winter, even when corrected for thermal inputs (accumulated degree days). Further, soil chemical parameters, microbial community structure, and fungal gene abundances remained altered at the end of 1 year, suggesting longer-term impacts on soil ecosystems beyond the initial pulse of decomposition products.
Keywords: biogeochemical hotspot; forensic taphonomy; human decomposition; necrobiome; soil biogeochemistry; soil microbiology.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures




Similar articles
-
From carrion to soil: microbial recycling of animal carcasses.Trends Microbiol. 2025 Feb;33(2):194-207. doi: 10.1016/j.tim.2024.09.003. Epub 2024 Oct 1. Trends Microbiol. 2025. PMID: 39358066 Review.
-
Soil pH amendment alters the abundance, diversity, and composition of microbial communities in two contrasting agricultural soils.Microbiol Spectr. 2024 Aug 6;12(8):e0416523. doi: 10.1128/spectrum.04165-23. Epub 2024 Jun 25. Microbiol Spectr. 2024. PMID: 38916324 Free PMC article.
-
Short-Term Transcriptional Response of Microbial Communities to Nitrogen Fertilization in a Pine Forest Soil.Appl Environ Microbiol. 2018 Jul 17;84(15):e00598-18. doi: 10.1128/AEM.00598-18. Print 2018 Aug 1. Appl Environ Microbiol. 2018. PMID: 29802185 Free PMC article.
-
Comparison of seasonal soil microbial process in snow-covered temperate ecosystems of northern China.PLoS One. 2014 Mar 25;9(3):e92985. doi: 10.1371/journal.pone.0092985. eCollection 2014. PLoS One. 2014. PMID: 24667929 Free PMC article.
-
Knowns and unknowns of the soil fungal necrobiome.Trends Microbiol. 2023 Feb;31(2):173-180. doi: 10.1016/j.tim.2022.08.011. Epub 2022 Sep 10. Trends Microbiol. 2023. PMID: 36100506 Review.
Cited by
-
Environmental predictors impact microbial-based postmortem interval (PMI) estimation models within human decomposition soils.PLoS One. 2024 Oct 11;19(10):e0311906. doi: 10.1371/journal.pone.0311906. eCollection 2024. PLoS One. 2024. PMID: 39392823 Free PMC article.
References
-
- Abarenkov K, Zirk A, Piirmann T et al. Full mothur UNITE+INSD dataset 1. Version 04.02.2020. Mothur, 2020.
-
- Apprill A, McNally S, Parsons R et al. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol. 2015;75:129–37. 10.3354/ame01753. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

