Cancer cell stiffening via CoQ10 and UBIAD1 regulates ECM signaling and ferroptosis in breast cancer
- PMID: 39294175
- PMCID: PMC11410950
- DOI: 10.1038/s41467-024-52523-y
Cancer cell stiffening via CoQ10 and UBIAD1 regulates ECM signaling and ferroptosis in breast cancer
Abstract
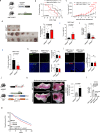
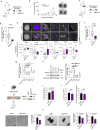
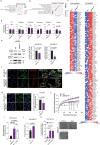
CoQ10 (Coenzyme Q10) is an essential fat-soluble metabolite that plays a key role in cellular metabolism. A less-known function of CoQ10 is whether it may act as a plasma membrane-stabilizing agent and whether this property can affect cancer development and progression. Here, we show that CoQ10 and its biosynthetic enzyme UBIAD1 play a critical role in plasmamembrane mechanical properties that are of interest for breast cancer (BC) progression and treatment. CoQ10 and UBIAD1 increase membrane fluidity leading to increased cell stiffness in BC. Furthermore, CoQ10 and UBIAD1 states impair ECM (extracellular matrix)-mediated oncogenic signaling and reduce ferroptosis resistance in BC settings. Analyses on human patients and mouse models reveal that UBIAD1 loss is associated with BC development and progression and UBIAD1 expression in BC limits CTCs (circulating tumor cells) survival and lung metastasis formation. Overall, this study reveals that CoQ10 and UBIAD1 can be further investigated to develop therapeutic interventions to treat BC patients with poor prognosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Morton, R. A. Ubiquinone. Nature182, 1764–1767 (1958). - PubMed
-
- Crane, F. L. Discovery of ubiquinone (coenzyme Q) and an overview of function. Mitochondrion7, S2–S7 (2007). - PubMed
-
- Navas, P., Villalba, J. M. & de Cabo, R. The importance of plasma membrane coenzyme Q in aging and stress responses. Mitochondrion7, S34–S40 (2007). - PubMed
-
- Agmo Hernández, V., Eriksson, E. K. & Edwards, K. Ubiquinone-10 alters mechanical properties and increases stability of phospholipid membranes. Biochim. Biophys. Acta (BBA) - Biomembr.1848, 2233–2243 (2015). - PubMed
-
- Sévin, D. C. & Sauer, U. Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat. Chem. Biol.10, 266–272 (2014). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- 20119/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- GGP20003/Fondazione Telethon (Telethon Foundation)
- 647057/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 963865/EC | EC Seventh Framework Programm | FP7 Ideas: European Research Council (FP7-IDEAS-ERC - Specific Programme: "Ideas" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

