Temporal BMP4 effects on mouse embryonic and extraembryonic development
- PMID: 39294373
- PMCID: PMC11485214
- DOI: 10.1038/s41586-024-07937-5
Temporal BMP4 effects on mouse embryonic and extraembryonic development
Abstract
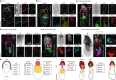
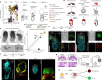
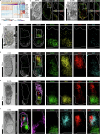
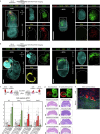
The developing placenta, which in mice originates through the extraembryonic ectoderm (ExE), is essential for mammalian embryonic development. Yet unbiased characterization of the differentiation dynamics of the ExE and its interactions with the embryo proper remains incomplete. Here we develop a temporal single-cell model of mouse gastrulation that maps continuous and parallel differentiation in embryonic and extraembryonic lineages. This is matched with a three-way perturbation approach to target signalling from the embryo proper, the ExE alone, or both. We show that ExE specification involves early spatial and transcriptional bifurcation of uncommitted ectoplacental cone cells and chorion progenitors. Early BMP4 signalling from chorion progenitors is required for proper differentiation of uncommitted ectoplacental cone cells and later for their specification towards trophoblast giant cells. We also find biphasic regulation by BMP4 in the embryo. The early ExE-originating BMP4 signal is necessary for proper mesoendoderm bifurcation and for allantois and primordial germ cell specification. However, commencing at embryonic day 7.5, embryo-derived BMP4 restricts the primordial germ cell pool size by favouring differentiation of their extraembryonic mesoderm precursors towards an allantois fate. ExE and embryonic tissues are therefore entangled in time, space and signalling axes, highlighting the importance of their integrated understanding and modelling in vivo and in vitro.
© 2024. The Author(s).
Conflict of interest statement
M.B.E. is a co-founder, scientific advisory board member, or consultant at TeraCyte, Primordium Labs, Spatial Genomics, and Asymptote Genetic Medicines. J.H.H. is an inventor on patents and patent applications related to ex utero embryogenesis, and a co-founder and chief scientific advisor of Renewal Bio, which has licensed the latter technologies. The other authors declare no competing interests.
Figures













References
-
- Rai, A. & Cross, J. C. Development of the hemochorial maternal vascular spaces in the placenta through endothelial and vasculogenic mimicry. Dev. Biol.387, 131–141 (2014). - PubMed
-
- Rossant, J. & Tam, P. P. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development136, 701–713 (2009). - PubMed
-
- Simmons, D. G. & Cross, J. C. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev. Biol.284, 12–24 (2005). - PubMed
-
- Arnold, S. J. & Robertson, E. J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol.10, 91–103 (2009). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

