Commensal consortia decolonize Enterobacteriaceae via ecological control
- PMID: 39294375
- PMCID: PMC11424487
- DOI: 10.1038/s41586-024-07960-6
Commensal consortia decolonize Enterobacteriaceae via ecological control
Abstract
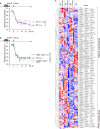
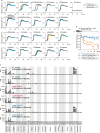
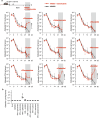
Persistent colonization and outgrowth of potentially pathogenic organisms in the intestine can result from long-term antibiotic use or inflammatory conditions, and may perpetuate dysregulated immunity and tissue damage1,2. Gram-negative Enterobacteriaceae gut pathobionts are particularly recalcitrant to conventional antibiotic treatment3,4, although an emerging body of evidence suggests that manipulation of the commensal microbiota may be a practical alternative therapeutic strategy5-7. Here we isolated and down-selected commensal bacterial consortia from stool samples from healthy humans that could strongly and specifically suppress intestinal Enterobacteriaceae. One of the elaborated consortia, comprising 18 commensal strains, effectively controlled ecological niches by regulating gluconate availability, thereby re-establishing colonization resistance and alleviating Klebsiella- and Escherichia-driven intestinal inflammation in mice. Harnessing these activities in the form of live bacterial therapies may represent a promising solution to combat the growing threat of proinflammatory, antimicrobial-resistant Enterobacteriaceae infection.
© 2024. The Author(s).
Conflict of interest statement
K.H. is a scientific advisory board member of Vedanta Biosciences and 4BIO CAPITAL. R.J.X. is co-founder of Jnana Therapeutics and Celsius Therapeutics, scientific advisory board member at Nestlé, and board director at MoonLake Immunotherapeutics. Y.A., M.U., K. Amafuji and Y.K. are employees of JSR corporation. J.M.N. and B.O. are employees of Vedanta Biosciences. S.C. was an employee of Vedanta Biosciences at the time of her contributions. D.R.P. is currently an employee of Novonesis. K.H., M.F., N.H., S.S., K. Atarashi, T.O. and Y.A. have filed international patent application PCT/JP2024/008014. The other authors declare no competing interests.
Figures













Update of
-
Rationally-defined microbial consortia suppress multidrug-resistant proinflammatory Enterobacteriaceae via ecological control.Res Sq [Preprint]. 2023 Oct 23:rs.3.rs-3462622. doi: 10.21203/rs.3.rs-3462622/v1. Res Sq. 2023. Update in: Nature. 2024 Sep;633(8031):878-886. doi: 10.1038/s41586-024-07960-6. PMID: 37961431 Free PMC article. Updated. Preprint.
References
-
- Antibiotic Resistance Threats in the United States, 2019 (Centers for Disease Control and Prevention, 2019); https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-re....
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

