Two-factor authentication underpins the precision of the piRNA pathway
- PMID: 39294378
- PMCID: PMC11499256
- DOI: 10.1038/s41586-024-07963-3
Two-factor authentication underpins the precision of the piRNA pathway
Abstract
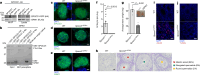
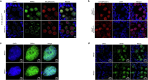
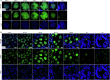
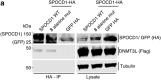
The PIWI-interacting RNA (piRNA) pathway guides the DNA methylation of young, active transposons during germline development in male mice1. piRNAs tether the PIWI protein MIWI2 (PIWIL4) to the nascent transposon transcript, resulting in DNA methylation through SPOCD1 (refs. 2-5). Transposon methylation requires great precision: every copy needs to be methylated but off-target methylation must be avoided. However, the underlying mechanisms that ensure this precision remain unknown. Here, we show that SPOCD1 interacts directly with SPIN1 (SPINDLIN1), a chromatin reader that primarily binds to H3K4me3-K9me3 (ref. 6). The prevailing assumption is that all the molecular events required for piRNA-directed DNA methylation occur after the engagement of MIWI2. We find that SPIN1 expression precedes that of both SPOCD1 and MIWI2. Furthermore, we demonstrate that young LINE1 copies, but not old ones, are marked by H3K4me3, H3K9me3 and SPIN1 before the initiation of piRNA-directed DNA methylation. We generated a Spocd1 separation-of-function allele in the mouse that encodes a SPOCD1 variant that no longer interacts with SPIN1. We found that the interaction between SPOCD1 and SPIN1 is essential for spermatogenesis and piRNA-directed DNA methylation of young LINE1 elements. We propose that piRNA-directed LINE1 DNA methylation requires a developmentally timed two-factor authentication process. The first authentication is the recruitment of SPIN1-SPOCD1 to the young LINE1 promoter, and the second is MIWI2 engagement with the nascent transcript. In summary, independent authentication events underpin the precision of piRNA-directed LINE1 DNA methylation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Ozata, D. M., Gainetdinov, I., Zoch, A., O’Carroll, D. & Zamore, P. D. PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet.20, 89–108 (2019). - PubMed
-
- De Fazio, S. et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature480, 259–263 (2011). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

