miRNA-mediated control of gephyrin synthesis drives sustained inhibitory synaptic plasticity
- PMID: 39294503
- PMCID: PMC11549329
- DOI: 10.1038/s44319-024-00253-z
miRNA-mediated control of gephyrin synthesis drives sustained inhibitory synaptic plasticity
Abstract
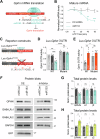
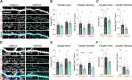
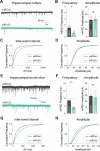
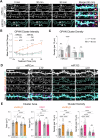
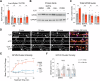
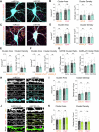
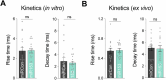
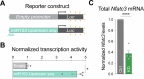
Activity-dependent protein synthesis is crucial for long-lasting forms of synaptic plasticity. However, our understanding of translational mechanisms controlling GABAergic synapses is limited. One distinct form of inhibitory long-term potentiation (iLTP) enhances postsynaptic clusters of GABAARs and the primary inhibitory scaffold, gephyrin, to promote sustained synaptic strengthening. While we previously found that persistent iLTP requires mRNA translation, the mechanisms controlling plasticity-induced gephyrin translation remain unknown. We identify miR153 as a novel regulator of Gphn mRNA translation which controls gephyrin protein levels and synaptic clustering, ultimately impacting inhibitory synaptic structure and function. iLTP induction downregulates miR153, reversing its translational suppression of Gphn mRNA and promoting de novo gephyrin protein synthesis and synaptic clustering during iLTP. Finally, we find that reduced miR153 expression during iLTP is driven by an excitation-transcription coupling pathway involving calcineurin, NFAT and HDACs, which also controls the miRNA-dependent upregulation of GABAARs. Together, we delineate a miRNA-dependent post-transcriptional mechanism that controls the expression of the key synaptic scaffold, gephyrin, and may converge with parallel miRNA pathways to coordinate gene upregulation to maintain inhibitory synaptic plasticity.
Keywords: GABAA Receptor; Gephyrin; Inhibitory Synapse; Translation; miRNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
miRNA-mediated control of gephyrin synthesis drives sustained inhibitory synaptic plasticity.bioRxiv [Preprint]. 2023 Dec 12:2023.12.12.570420. doi: 10.1101/2023.12.12.570420. bioRxiv. 2023. Update in: EMBO Rep. 2024 Nov;25(11):5141-5168. doi: 10.1038/s44319-024-00253-z. PMID: 38168421 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01 MH119154/MH/NIMH NIH HHS/United States
- F31 NS130979/NS/NINDS NIH HHS/United States
- R35 NS116879/NS/NINDS NIH HHS/United States
- T32NS099042/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- F31NS130979/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01MH119154/HHS | NIH | National Institute of Mental Health (NIMH)
- R01 NS095809/NS/NINDS NIH HHS/United States
- F30DA048543/HHS | NIH | National Institute on Drug Abuse (NIDA)
- R01MH128199/HHS | NIH | National Institute of Mental Health (NIMH)
- R01 MH123700/MH/NIMH NIH HHS/United States
- T32 NS099042/NS/NINDS NIH HHS/United States
- R01NS95809/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- F99 NS120640/NS/NINDS NIH HHS/United States
- R01 MH128199/MH/NIMH NIH HHS/United States
- F31NS12410/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01DA35821/HHS | NIH | National Institute on Drug Abuse (NIDA)
- R35NS116879/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01 DA035821/DA/NIDA NIH HHS/United States
- F30 DA048543/DA/NIDA NIH HHS/United States
- FNS120640A/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
LinkOut - more resources
Full Text Sources

