Screening and identification of susceptibility genes for cervical cancer via bioinformatics analysis and the construction of an mitophagy-related genes diagnostic model
- PMID: 39294534
- PMCID: PMC11410911
- DOI: 10.1007/s00432-024-05952-7
Screening and identification of susceptibility genes for cervical cancer via bioinformatics analysis and the construction of an mitophagy-related genes diagnostic model
Retraction in
-
Retraction Note: Screening and identification of susceptibility genes for cervical cancer via bioinformatics analysis and the construction of an mitophagy-related genes diagnostic model.J Cancer Res Clin Oncol. 2025 Jan 23;151(1):42. doi: 10.1007/s00432-025-06100-5. J Cancer Res Clin Oncol. 2025. PMID: 39843805 Free PMC article. No abstract available.
Abstract
Purpose: This study aims to utilize bioinformatics methods to systematically screen and identify susceptibility genes for cervical cancer, as well as to construct and validate an mitophagy-related genes (MRGs) diagnostic model. The objective is to increase the understanding of the disease's pathogenesis and improve early diagnosis and treatment.
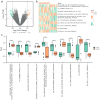
Method: We initially collected a large amount of genomic data, including gene expression profile and single nucleotide polymorphism (SNP) data, from the control group and Cervical cancer (CC) patients. Through bioinformatics analysis, which employs methods such as differential gene expression analysis and pathway enrichment analysis, we identified a set of candidate susceptibility genes associated with cervical cancer.
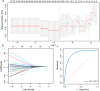
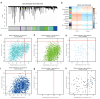
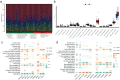
Results: MRGs were extracted from single-cell RNA sequencing data, and a network graph was constructed on the basis of intercellular interaction data. Furthermore, using machine learning algorithms, we constructed a clinical prognostic model and validated and optimized it via extensive clinical data. Through bioinformatics analysis, we successfully identified a group of genes whose expression significantly differed during the development of CC and revealed the biological pathways in which these genes are involved. Moreover, our constructed clinical prognostic model demonstrated excellent performance in the validation phase, accurately predicting the clinical prognosis of patients.
Conclusion: This study delves into the susceptibility genes of cervical cancer through bioinformatics approaches and successfully builds a reliable clinical prognostic model. This study not only helps uncover potential pathogenic mechanisms of cervical cancer but also provides new directions for early diagnosis and treatment of the disease.
Keywords: Bioinformatics analysis; Cervical cancer; Clinical prognosis model; Susceptibility genes.
© 2024. The Author(s).
Conflict of interest statement
The author did not encounter any conflicts throughout the preparation of the article.
Figures