Low-dose pro-resolving mediators temporally reset the resolution response to microbial inflammation
- PMID: 39294573
- PMCID: PMC11411770
- DOI: 10.1186/s10020-024-00877-w
Low-dose pro-resolving mediators temporally reset the resolution response to microbial inflammation
Abstract
Background: Specialized pro-resolving mediators (SPMs) promote resolution of inflammation, clear infections and stimulate tissue regeneration. These include resolvins, protectins, and maresins. During self-resolving acute inflammation, SPMs are produced and have key functions activating endogenous resolution response for returning to homeostasis. Herein, we addressed whether infections initiated with ongoing inflammation alter resolution programs, and if low-dose repetitive SPM regimen re-programs the resolution response.
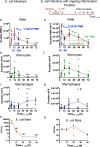
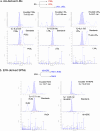
Methods: Inflammation was initiated with zymosan (1 mg/mouse) followed by E. coli (105 CFU/mouse) infections carried out in murine peritonitis, and exudates collected at 4-72 h. Leukocytes were enumerated using light microscopy, percentages of PMN, monocytes and macrophages were determined using flow cytometry, and resolution indices calculated. Lipid mediators and SPM profiles were established using mass spectrometry-based metabololipidomics. Repetitive dosing with a SPM panel consisting of RvD1, RvD2, RvD5, MaR1 and RvE2 (0.1 ng/mouse each, i.p.) was given to mice, followed by zymosan challenge. Leukocyte composition, resolution indices and RNA-sequencing were carried out for the repetitive SPM treatments.
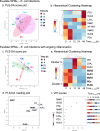
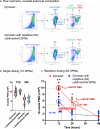
Results: E. coli infections initiated acute inflammation-resolution programs with temporal SPM production in the infectious exudates. Zymosan-induced inflammation prior to E. coli peritonitis shifted exudate resolution indices and delayed E. coli clearance. Lipid mediator metabololipidomics demonstrated that E. coli infection with ongoing zymosan-induced inflammation shifted the time course of exudate SPMs, activating a SPM cluster that included RvD1, RvD5 and MaR1 during the initiation phase of infectious inflammation (0-4 h); RvD5 and MaR1 were present also in the resolution phase (24-48 h). To emulate daily SPM regimens used in humans, a repetitive subthreshold dosing of the SPM panel RvD1, RvD2, RvD5, MaR1 and RvE2 each at 0.1 ng per mouse was administered. This low-dose SPM regimen accelerated exudate PMN clearance following zymosan-induced inflammation, and shortened the resolution interval by > 70%. These low-dose SPMs regulated genes and pathways related to immune response, chemokine clearance and tissue repair, as demonstrated by using RNA-sequencing.
Conclusions: Infections encountered during ongoing inflammation in mice reset the resolution mechanisms of inflammation via SPM clusters. Low-dose SPMs activate innate immune responses and pathways towards the resolution response that can be reprogrammed.
Keywords: LC–MS–MS; Macrophage; Maresins; Neutrophil; Protectins; Resolvins; SPMs.
© 2024. The Author(s).
Conflict of interest statement
The authors declared that there are no competing interests.
Figures


Similar articles
-
Infection regulates pro-resolving mediators that lower antibiotic requirements.Nature. 2012 Apr 25;484(7395):524-8. doi: 10.1038/nature11042. Nature. 2012. PMID: 22538616 Free PMC article.
-
E-series resolvin metabolome, biosynthesis and critical role of stereochemistry of specialized pro-resolving mediators (SPMs) in inflammation-resolution: Preparing SPMs for long COVID-19, human clinical trials, and targeted precision nutrition.Semin Immunol. 2022 Jan;59:101597. doi: 10.1016/j.smim.2022.101597. Epub 2022 Feb 16. Semin Immunol. 2022. PMID: 35227568 Free PMC article. Review.
-
Plasticity of leukocytic exudates in resolving acute inflammation is regulated by MicroRNA and proresolving mediators.Immunity. 2013 Nov 14;39(5):885-98. doi: 10.1016/j.immuni.2013.10.011. Immunity. 2013. PMID: 24238341 Free PMC article.
-
Human milk proresolving mediators stimulate resolution of acute inflammation.Mucosal Immunol. 2016 May;9(3):757-766. doi: 10.1038/mi.2015.99. Epub 2015 Oct 14. Mucosal Immunol. 2016. PMID: 26462421 Free PMC article.
-
Novel mediators and mechanisms in the resolution of infectious inflammation: evidence for vagus regulation.J Intern Med. 2019 Sep;286(3):240-258. doi: 10.1111/joim.12871. Epub 2019 Jan 18. J Intern Med. 2019. PMID: 30565762 Review.
Cited by
-
Oxylipins as canine sepsis indicators in vivo and in ex vivo skin organ culture model.Sci Rep. 2025 Apr 11;15(1):12483. doi: 10.1038/s41598-025-97460-y. Sci Rep. 2025. PMID: 40216925 Free PMC article.
References
-
- Bardin M, Pawelzik SC, Lagrange J, Mahdi A, Arnardottir H, Regnault V, et al. The resolvin D2 - GPR18 axis is expressed in human coronary atherosclerosis and transduces atheroprotection in apolipoprotein E deficient mice. Biochem Pharmacol. 2022;201: 115075. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

