Transcriptomic profiling reveals the complex interaction between a bipartite begomovirus and a cucurbitaceous host plant
- PMID: 39294575
- PMCID: PMC11409788
- DOI: 10.1186/s12864-024-10781-6
Transcriptomic profiling reveals the complex interaction between a bipartite begomovirus and a cucurbitaceous host plant
Abstract
Background: Begomoviruses are major constraint in the production of many crops. Upon infection, begomoviruses may substantially modulate plant biological processes. While how monopartite begomoviruses interact with their plant hosts has been investigated extensively, bipartite begomoviruses-plant interactions are understudied. Moreover, as one of the major groups of hosts, cucurbitaceous plants have been seldom examined in the interaction with begomoviruses.
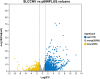
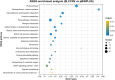
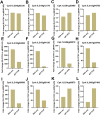
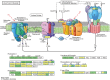
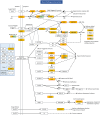
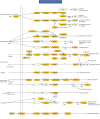
Results: We profiled the zucchini transcriptomic changes induced by a bipartite begomovirus squash leaf curl China virus (SLCCNV). We identified 2275 differentially-expressed genes (DEGs), of which 1310 were upregulated and 965 were downregulated. KEGG enrichment analysis of the DEGs revealed that many pathways related to primary and secondary metabolisms were enriched. qRT-PCR verified the transcriptional changes of twelve selected DEGs induced by SLCCNV infection. Close examination revealed that the expression levels of all the DEGs of the pathway Photosynthesis were downregulated upon SLCCNV infection. Most DEGs in the pathway Plant-pathogen interaction were upregulated, including some positive regulators of plant defenses. Moreover, the majority of DEGs in the MAPK signaling pathway-plant were upregulated.
Conclusion: Our findings indicates that SLCCNV actively interact with its cucurbitaceous plant host by suppressing the conversion of light energy to chemical energy and inducing immune responses. Our study not only provides new insights into the interactions between begomoviruses and host plants, but also adds to our knowledge on virus-plant interactions in general.
Keywords: Cucurbita pepo; Bipartite begomovirus; Plant-virus interaction; Squash leaf curl China virus; Transcriptional reprograming.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Osterbaan LJ, Fuchs M. Dynamic interactions between plant viruses and their hosts for symptom development. J Plant Pathol. 2019;101:885–95. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

