iPLA2β loss leads to age-related cognitive decline and neuroinflammation by disrupting neuronal mitophagy
- PMID: 39294744
- PMCID: PMC11409585
- DOI: 10.1186/s12974-024-03219-z
iPLA2β loss leads to age-related cognitive decline and neuroinflammation by disrupting neuronal mitophagy
Abstract
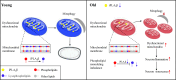
Background: During brain aging, disturbances in neuronal phospholipid metabolism result in impaired cognitive function and dysregulation of neurological processes. Mutations in iPLA2β are associated with neurodegenerative conditions that significantly impact brain phospholipids. iPLA2β deficiency exacerbates mitochondrial dysfunction and abnormal mitochondrial accumulation. We hypothesized that iPLA2β contributes to age-related cognitive decline by disrupting neuronal mitophagy.
Methodology: We used aged wild-type (WT) mice and iPLA2β-/- mice as natural aging models to assess cognitive performance, iPLA2β expression in the cortex, levels of chemokines and inflammatory cytokines, and mitochondrial dysfunction, with a specific focus on mitophagy and the mitochondrial phospholipid profile. To further elucidate the role of iPLA2β, we employed adeno-associated virus (AAV)-mediated iPLA2β overexpression in aged mice and re-evaluated these parameters.
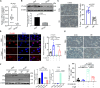
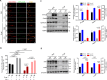
Results: Our findings revealed a significant reduction in iPLA2β levels in the prefrontal cortex of aged brains. Notably, iPLA2β-deficient mice exhibited impaired learning and memory. Loss of iPLA2β in the PFC of aged mice led to increased levels of chemokines and inflammatory cytokines. This damage was associated with altered mitochondrial morphology, reduced ATP levels due to dysregulation of the parkin-independent mitophagy pathway, and changes in the mitochondrial phospholipid profile. AAV-mediated overexpression of iPLA2β alleviated age-related parkin-independent mitophagy pathway dysregulation in primary neurons and the PFC of aged mice, reduced inflammation, and improved cognitive function.
Conclusions: Our study suggests that age-related iPLA2β loss in the PFC leads to cognitive decline through the disruption of mitophagy. These findings highlight the potential of targeting iPLA2β to ameliorate age-related neurocognitive disorders.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
- 82101640/the National Natural Science Foundation of China
- 2021-I2M-1-024/CAMS Innovation Fund for Medical Sciences
- 2019-RC-HL-017/Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences
- 3332018127/Peking Union Medical College Central University Basic Research Funding
- 2022YFA1103803/the National Key R&D Program of China
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

